/*******Cerebellar Golgi Cell Model with Na+/K+ ATPase**********
Developers: Fabio M Simoes de Souza & E De Schutter
Work Progress: July 2009 - Dec 2009
Developed At: Okinawa Institute of Science and Technology
Computational Neuroscience Unit
Okinawa - Japan
Model Published in:
Botta P, Simoes de Souza F, Sangrey T, De Schutter E,
Valenzuela F (2010) Alcohol excites cerebellar Golgi
cells by inhibiting the Na+/K+ ATPase.
Neuropsychopharmacology 35: 1984-1996.
This script is a modification from a previous published GoC model
(Solinas et al., 2007). A Na+/K+ ATPase and ionic concentration pools
for Na+, K+, Ca2+ were incorporated into the soma of the model.
The equations that simulated the Na+/K+ ATPase are described in Table
S10 of Takeuchi et al. (2006)
Abstract
Alcohol Excites Cerebellar Golgi Cells by Inhibiting the Na+/K+ ATPase
Patch-clamp in cerebellar slices and computer modeling show that
ethanol excites Golgi cells by inhibiting the Na+/K+ ATPase. In
particular, voltage-clamp recordings of Na+/K+ ATPase currents
indicated that ethanol partially inhibits this pump and this effect
could be mimicked by low concentrations of the Na+/K+ ATPase blocker
ouabain. The partial inhibition of Na+/K+ ATPase in a computer model
of the Golgi cell reproduced these experimental findings that
established a novel mechanism of action of ethanol on neural
excitability.
Reference: Botta P, de Souza FM, Sangrey T, De Schutter E, Valenzuela
CF (2010) Alcohol excites cerebellar Golgi cells by inhibiting the
na(+)-k(+) ATPase. Neuropsychopharmacology 35:1984-96
Usage instructions:
Auto-launch from ModelDB or download and extract the archive. Then
under:
----
MSWIN
run mknrndll, cd to the archive and make the nrnmech.dll. Then double
click on the mosinit.hoc file.
----
MAC OS X
Drag and drop the Golgi_cell_NaKATPAse folder onto the mknrndll icon.
Drag and drop the mosinit.hoc file onto the nrngui icon.
----
Linux/Unix
Change directory to the Golgi_cell_NaKATPAse folder. run
nrnivmodl. Then type nrngui mosinit.hoc
----
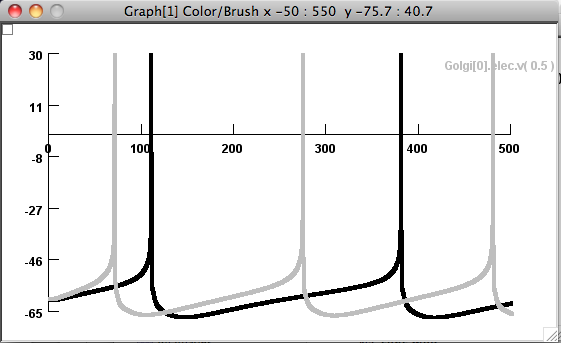
Once the simulation is started click the button labeled
"Figure 7g from Botta et al. 2010" The simulation will produce a
result similar to figure 7g from the paper:
 20140305 An update was made to pump.mod to make the model current with
a new version of NEURON (hg revision 1027) while retaining
compatibility with old versions. The number of parameters in settables
was made to match the number of the calling instances.
Changelog
--------
* 20220924: Update MOD files to avoid declaring variables and functions with the same name.
See https://github.com/neuronsimulator/nrn/pull/1992
20140305 An update was made to pump.mod to make the model current with
a new version of NEURON (hg revision 1027) while retaining
compatibility with old versions. The number of parameters in settables
was made to match the number of the calling instances.
Changelog
--------
* 20220924: Update MOD files to avoid declaring variables and functions with the same name.
See https://github.com/neuronsimulator/nrn/pull/1992
 20140305 An update was made to pump.mod to make the model current with
a new version of NEURON (hg revision 1027) while retaining
compatibility with old versions. The number of parameters in settables
was made to match the number of the calling instances.
Changelog
--------
* 20220924: Update MOD files to avoid declaring variables and functions with the same name.
See https://github.com/neuronsimulator/nrn/pull/1992
20140305 An update was made to pump.mod to make the model current with
a new version of NEURON (hg revision 1027) while retaining
compatibility with old versions. The number of parameters in settables
was made to match the number of the calling instances.
Changelog
--------
* 20220924: Update MOD files to avoid declaring variables and functions with the same name.
See https://github.com/neuronsimulator/nrn/pull/1992