Model files for the entry "Olfactory Computations in Mitral-Granule
Cell Circuits" of the Springer Encyclopedia of Computational
Neuroscience by Michele Migliore and Tom Mctavish.
The simulations illustrate two typical Mitral-Granule cell circuits in
the olfactory bulb of vertebrates.
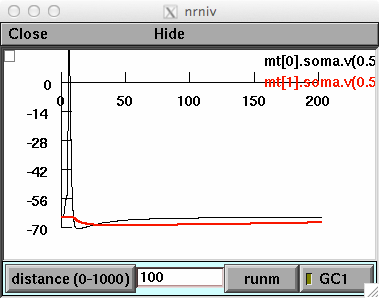
The simulation file forfig1-springer.hoc refers to Fig.1 of the entry,
and shows the somatic membrane potential of the two mitral cells.
A somatic AP is elicited in M0 with a short current pulse, and
generates an inhibitory postsynaptic potential in M1 with a peak
amplitude that depends on the relative distance between the soma of
the two mitral cells, unless GC1 is active. Users can change the
active state of GC1 and the distance between M0 and M1, to see the
distance-independence of the lateral inhibition when GC1 is active.
Here is what you should see after running the forfig1-springer.hoc
model in its default configuration:
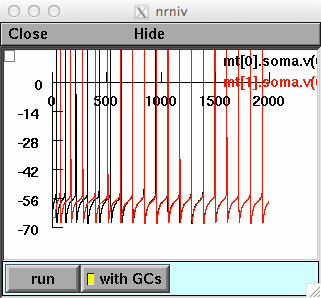
 forfig2.hoc reproduces the traces shown in Fig.2. The same somatic
current injection (0.15nA) is delivered to both Mitral cells, with an
initial relative latency. When the GCs are activated, at t=500, the
firing activity will be synchronized:
forfig2.hoc reproduces the traces shown in Fig.2. The same somatic
current injection (0.15nA) is delivered to both Mitral cells, with an
initial relative latency. When the GCs are activated, at t=500, the
firing activity will be synchronized:
 forfig3-springer.hoc reproduces Fig.3 of the entry, and illustrate a
gating effect:
forfig3-springer.hoc reproduces Fig.3 of the entry, and illustrate a
gating effect:
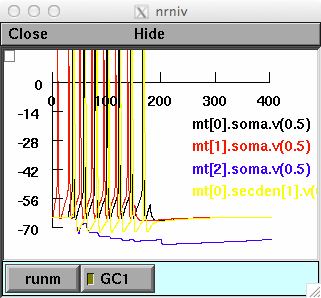 The plots show membrane potential of M0-M2 soma and lateral dendrite
of M0 at 800um from the soma, and the M0 membrane potential as a
function of distance from the soma.
In the simulations, all mitral cells receive the same input. Note
that firing of M2 depends on the activation of GC1 by M1, which gates
the backpropagation of the APs along the M0 lateral dendrite. As a
consequence, GC2 will not be activated and M2 can fire APs. With GC1
inactive, APs from M0 backpropagate until they activate GC2,
inhibiting M2.
Under unix systems:
to compile the mod files use the command
nrnivmodl
and run the simulation file with the command
nrngui filename
Under Windows systems:
to compile the mod files use the "mknrndll" command.
A double click on a simulation file
will open the simulation window.
Questions on how to use this model
should be directed to michele.migliore@cnr.it
The plots show membrane potential of M0-M2 soma and lateral dendrite
of M0 at 800um from the soma, and the M0 membrane potential as a
function of distance from the soma.
In the simulations, all mitral cells receive the same input. Note
that firing of M2 depends on the activation of GC1 by M1, which gates
the backpropagation of the APs along the M0 lateral dendrite. As a
consequence, GC2 will not be activated and M2 can fire APs. With GC1
inactive, APs from M0 backpropagate until they activate GC2,
inhibiting M2.
Under unix systems:
to compile the mod files use the command
nrnivmodl
and run the simulation file with the command
nrngui filename
Under Windows systems:
to compile the mod files use the "mknrndll" command.
A double click on a simulation file
will open the simulation window.
Questions on how to use this model
should be directed to michele.migliore@cnr.it
 forfig2.hoc reproduces the traces shown in Fig.2. The same somatic
current injection (0.15nA) is delivered to both Mitral cells, with an
initial relative latency. When the GCs are activated, at t=500, the
firing activity will be synchronized:
forfig2.hoc reproduces the traces shown in Fig.2. The same somatic
current injection (0.15nA) is delivered to both Mitral cells, with an
initial relative latency. When the GCs are activated, at t=500, the
firing activity will be synchronized:
 forfig3-springer.hoc reproduces Fig.3 of the entry, and illustrate a
gating effect:
forfig3-springer.hoc reproduces Fig.3 of the entry, and illustrate a
gating effect:
 The plots show membrane potential of M0-M2 soma and lateral dendrite
of M0 at 800um from the soma, and the M0 membrane potential as a
function of distance from the soma.
In the simulations, all mitral cells receive the same input. Note
that firing of M2 depends on the activation of GC1 by M1, which gates
the backpropagation of the APs along the M0 lateral dendrite. As a
consequence, GC2 will not be activated and M2 can fire APs. With GC1
inactive, APs from M0 backpropagate until they activate GC2,
inhibiting M2.
Under unix systems:
to compile the mod files use the command
nrnivmodl
and run the simulation file with the command
nrngui filename
Under Windows systems:
to compile the mod files use the "mknrndll" command.
A double click on a simulation file
will open the simulation window.
Questions on how to use this model
should be directed to michele.migliore@cnr.it
The plots show membrane potential of M0-M2 soma and lateral dendrite
of M0 at 800um from the soma, and the M0 membrane potential as a
function of distance from the soma.
In the simulations, all mitral cells receive the same input. Note
that firing of M2 depends on the activation of GC1 by M1, which gates
the backpropagation of the APs along the M0 lateral dendrite. As a
consequence, GC2 will not be activated and M2 can fire APs. With GC1
inactive, APs from M0 backpropagate until they activate GC2,
inhibiting M2.
Under unix systems:
to compile the mod files use the command
nrnivmodl
and run the simulation file with the command
nrngui filename
Under Windows systems:
to compile the mod files use the "mknrndll" command.
A double click on a simulation file
will open the simulation window.
Questions on how to use this model
should be directed to michele.migliore@cnr.it