Model files that accompany the publication:
Pezo, Soudry, Orio (2014) Front Comp Neurosci 8:139. DIFFUSION
APPROXIMATION-BASED SIMULATION OF STOCHASTIC ION CHANNELS: WHICH
METHOD TO USE?
DOI: 10.3389/fncom.2014.00139
Sample codes for stochastic simulation of ion channels in NEURON
HH folder:
----------
Original H&H model. Single compartment simulation, with the option of
a short simulation with voltage trace or a long simulation with
inter-spike intervals recording.
SchmidtHieber folder:
---------------------
Simplified hippocampal neuron with fast kinetic in sodium channels
(Schmidt-Hieber and Bischofberger (2010) J. Neurosci 30:10233.) For
this publication, inactivation of potassium channels was removed
In each case, several stochastic simulation algorithms are provided:
MC: Markov chain modeling, using Gillespie's algorithm
Gillespie (1977) Exact stochastic simulation of coupled chemical
reactions. J. Phys. Chem.81:2340
DA: (unbound) diffusion approximation
Orio and Soudry (2012) Simple, Fast and Accurate Implementation of the
Diffusion Approximation Algorithm for Stochastic Ion Channels with
Multiple States. PLOS ONE 7(5):e36670
SSmc: Stochastic Shielding approximation with Markov Chains for
stochastic transitions
Schmandt and Galan (2012) Stochastic-Shielding Approximation of Markov
Chains and its Application to Efficiently Simulate Random Ion-Channel
Gating. Phys Rev Lett 109:118101
SSda: Stochastic Shielding approximation with Diffusion approximation
in stochsatic terms
Combination of DA and SSmc, published here for the first time.
Ref: Diffusion Approximation with a reflection procedure to control
boundary and normalization constraints
Dangerfield et al. (2012) Modeling ion channel dynamics through
reflected stochastic differential equations. Physical Review E
85(5):051907.
TR: Diffusion Approximation with a Truncation/Restoration procedure to
control boundary and normalization constraints.
Huang et al. (2013) Phys Rev E 87:012716
CN: Uncoupled gating particles with Colored Noise (Only applied to
H&H)
Guler (2013) Stochastic Hodgkin-Huxley Equations with Colored Noise
Terms in the Conductances Neural Comp 25:46-74
Demo run: Autolaunch from ModelDB (after NEURON is installed and the
browser configured) or download this archive, expand it, and compile
the mod files in the HH folder (mknrndll on mswin or mac, nrnivmodl on
unix/linux), finally start by running mosinit.hoc (double click in
windows explorer, drag and drop onto nrngu on the mac, or type "nrngui
mosinit.hoc" on the command line in unix/linux). Once the simulation
is running click accept on the first window:
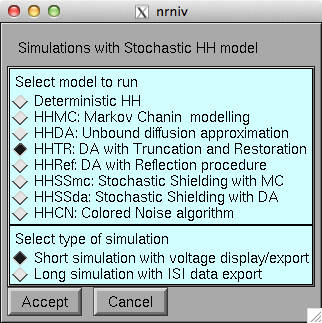 and run on the second to obtain a demo run like the below:
and run on the second to obtain a demo run like the below:
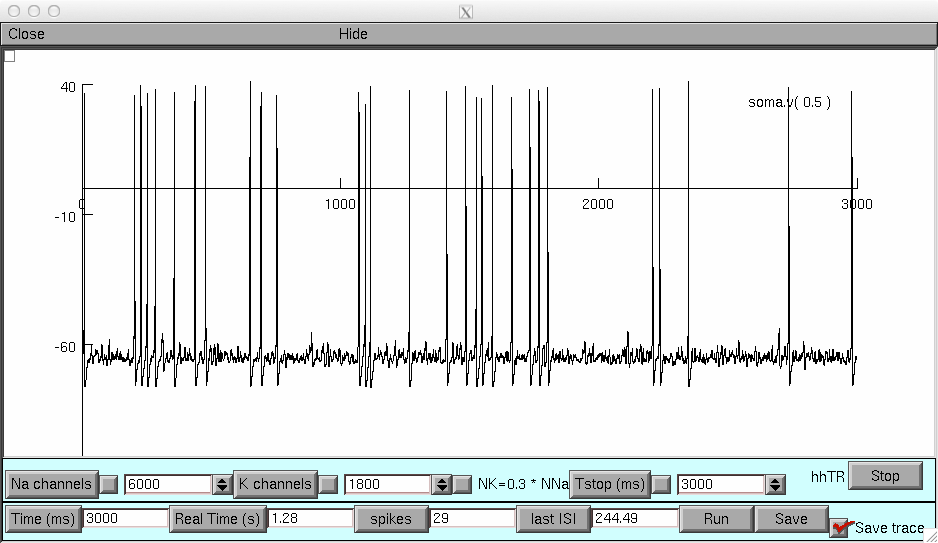
 and run on the second to obtain a demo run like the below:
and run on the second to obtain a demo run like the below:

and run on the second to obtain a demo run like the below:
