This code reproduces the simulations described in the paper
"Simulation neurotechnologies for advancing brain research:
Parallelizing large networks in NEURON", Lytton et al. 2016, Neural
Computation.
Two versions of the model are provided, one using NetPyNE and a
simplified version without using NetPyNE.
NetPyNE is a python package to facilitate the development, parallel
simulation and analysis of biological neuronal networks using the
NEURON simulator. More information: www.neurosimlab.org/netpyne
* NetPyNE version:
Three different networks are included: using only Hodgkin-Huxley cells
(HHNet.py), Izhikevich cells (IzhiNet.py), and a hybrid one with both
cell models (HybridNet.py). Note that for the Izhikevich net you will
need to compile the mod file via: nrniv izhi2007b.mod.
To test the simulation choose which parameter file to use in init.py
and run the file:
- single processor: python -i init.py
which should produce this image like figure 2 in the paper:
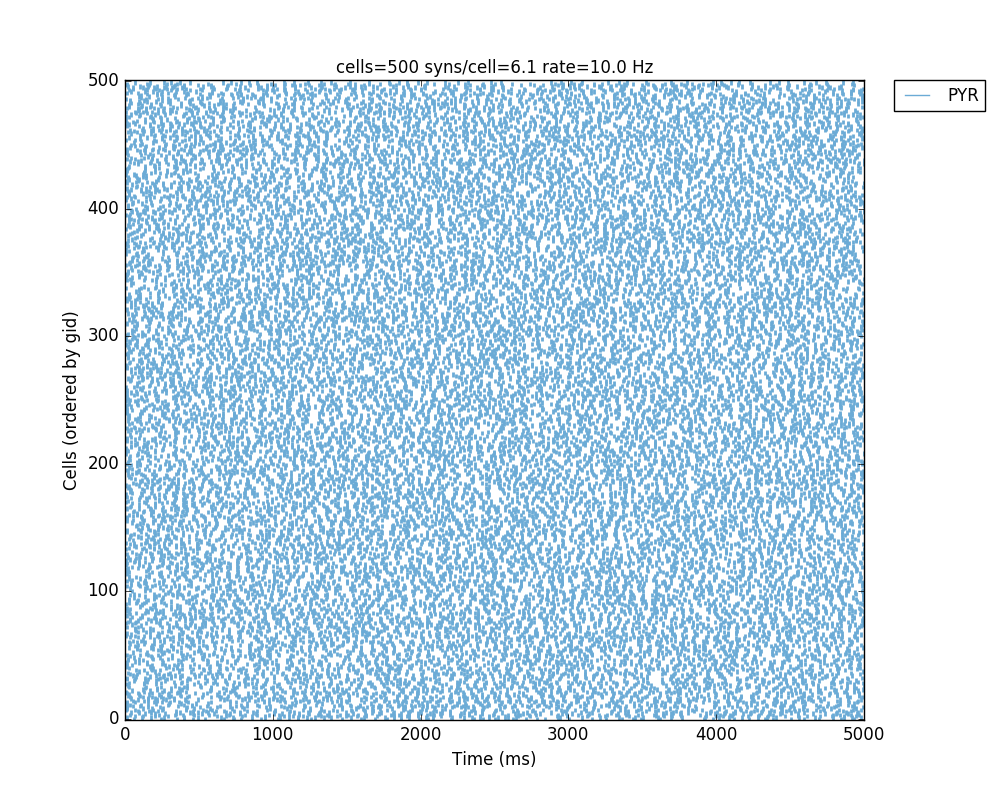 - in parallel: mpiexec -n 4 nrniv -mpi -python init.py
* Simplified version without NetPyNE:
This version only generates the network of Hodgkin-Huxley type
cells. To run the simulation, execute the file simplenet.py:
- single processor: python simplenet.py
- in parallel: mpiexec -n 4 nrniv -mpi -python simplenet.py
For questions/comments email: Salvador Dura-Bernal
(salvadordura at gmail.com) or Bill Lytton (billl at neurosim.downstate.edu)
- in parallel: mpiexec -n 4 nrniv -mpi -python init.py
* Simplified version without NetPyNE:
This version only generates the network of Hodgkin-Huxley type
cells. To run the simulation, execute the file simplenet.py:
- single processor: python simplenet.py
- in parallel: mpiexec -n 4 nrniv -mpi -python simplenet.py
For questions/comments email: Salvador Dura-Bernal
(salvadordura at gmail.com) or Bill Lytton (billl at neurosim.downstate.edu)
 - in parallel: mpiexec -n 4 nrniv -mpi -python init.py
* Simplified version without NetPyNE:
This version only generates the network of Hodgkin-Huxley type
cells. To run the simulation, execute the file simplenet.py:
- single processor: python simplenet.py
- in parallel: mpiexec -n 4 nrniv -mpi -python simplenet.py
For questions/comments email: Salvador Dura-Bernal
(salvadordura at gmail.com) or Bill Lytton (billl at neurosim.downstate.edu)
- in parallel: mpiexec -n 4 nrniv -mpi -python init.py
* Simplified version without NetPyNE:
This version only generates the network of Hodgkin-Huxley type
cells. To run the simulation, execute the file simplenet.py:
- single processor: python simplenet.py
- in parallel: mpiexec -n 4 nrniv -mpi -python simplenet.py
For questions/comments email: Salvador Dura-Bernal
(salvadordura at gmail.com) or Bill Lytton (billl at neurosim.downstate.edu)