Model files in the NEURON simulation environment (v7.5) from the paper
"The physiological
variability of channel density in hippocampal CA1 pyramidal cells
and interneurons explored using a unified
data-driven modeling workflow"
by Rosanna Migliore, Carmen
A. Lupascu, Luca L. Bologna, Armando Romani,
Jean-Denis Courcol,
Stefano Antonel, Werner A.H. Van Geit, Alex M. Thomson, Audrey Mercer, Sigrun Lange, Joanne Falck,
Christian A. Roessert, Ying Shi, Olivier Hagens, Maurizio Pezzoli, Tamas F. Freund, Szabolcs Kali,
Eilif B. Muller, Felix Schuermann, Henry Markram, and Michele Migliore
PLOS Computational Biology 2018
Usage:
Auto-launch from ModelDB or:
Compile the mod files with mknrndll (mswin or graphical mac) or
nrnivmodl (unix/linux)). Start the simulation by (unix/linux) typing
on the command line:
nrngui mosinit.hoc
or (mac os x) drag and dropping the mosinit.hoc file on the nrngui
icon or (mswin) double clicking on the mosinit.hoc file.
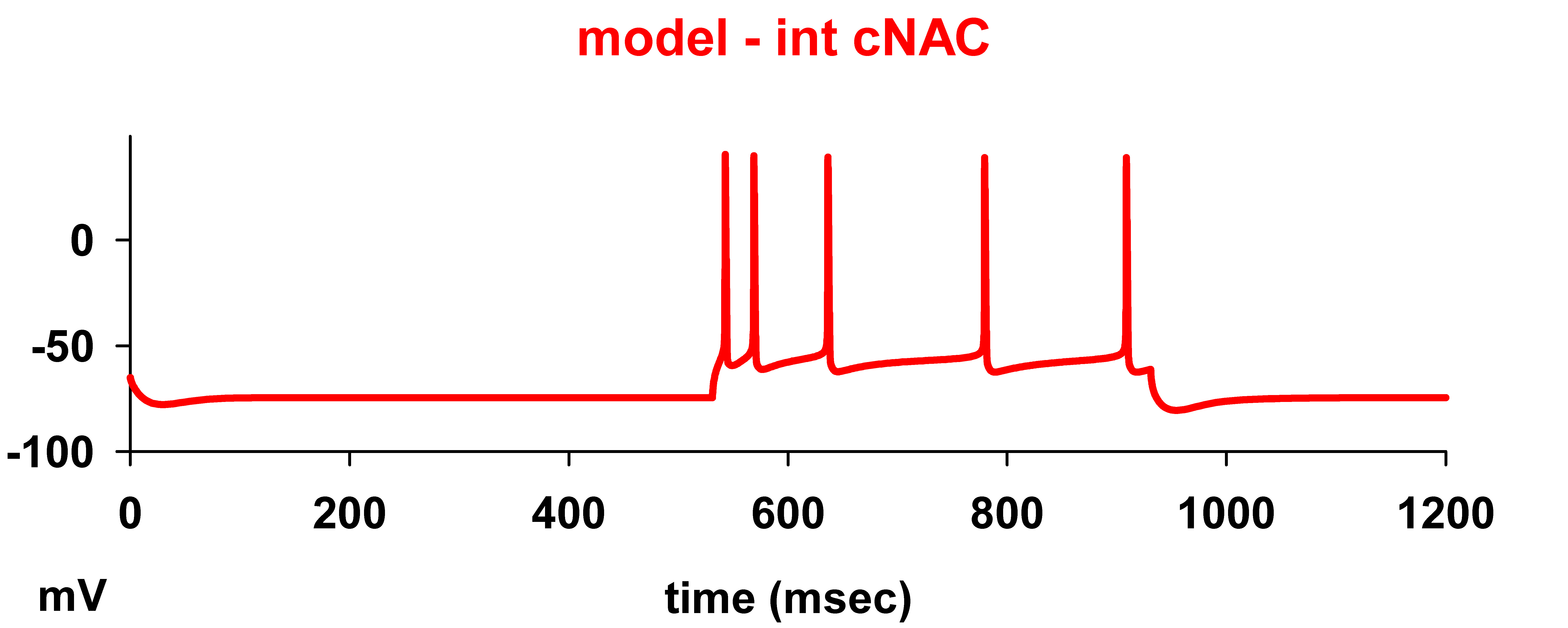
After selecting the e-type and the corresponding input current, simulation will reproduce the optimized somatic membrane potential as in fig4A (red traces) of the paper.
Example: clicking on run int cNAN 0.4 nA on the initial window should produce the following graph:
Questions on how to use this model should be directed to rosanna.migliore@cnr.it
All electrophysiological traces and morphologies used for the models
are available at the Human Brain Project collab https://collab.humanbrainproject.eu/#/collab/18565
Access to this resource requires free user registration at https://www.humanbrainproject.eu/en/hbp-platforms/getting-access/