Here are files associated with a modeling paper in Press in PLoS Comp
Biol.
This computer code was contributed by Avrama Blackwell
This is a multi-compartmental, stochastic version of the Kim et
al. 2010 paper. Here there are a few additional reactions, and some
of the rate constants have been updated. It addresses the role of
molecule anchoring in PKA dependent hippocampal LTP.
1. KimBlackwellPLoSCompBiol_NeuroRDxml.tar contains the xml for
default parameters and for the spine neck length variations.
2. StocMeshMorphMol.jar - java program for extracting concentration of
particular molecules averaged over different spatial regions
3. StocBatch_ABDE - batch file to run the java program to extract the
molecules graphed in the figures describing default simulations
(effect of location).
4. stochdiff-2.0.3-mol.jar - version of NeuroRD, which outputs
molecule quantity, used for the simulations. The output of this file
is processed by StocMeshMorphMol.
A single simulation takes several days to run.
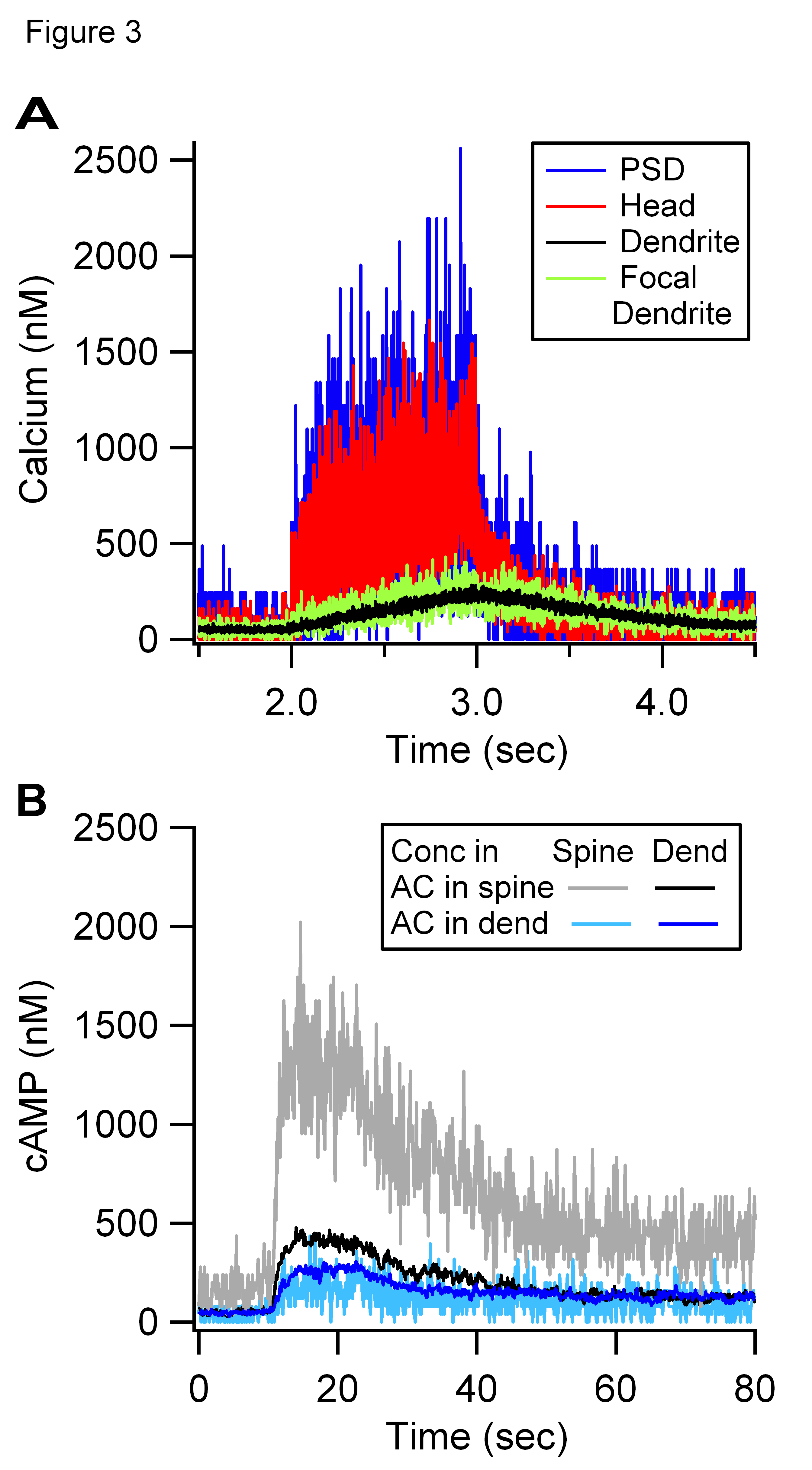 Above Figure 3 shows the output from two simulations.
Above Figure 3 shows the output from two simulations.
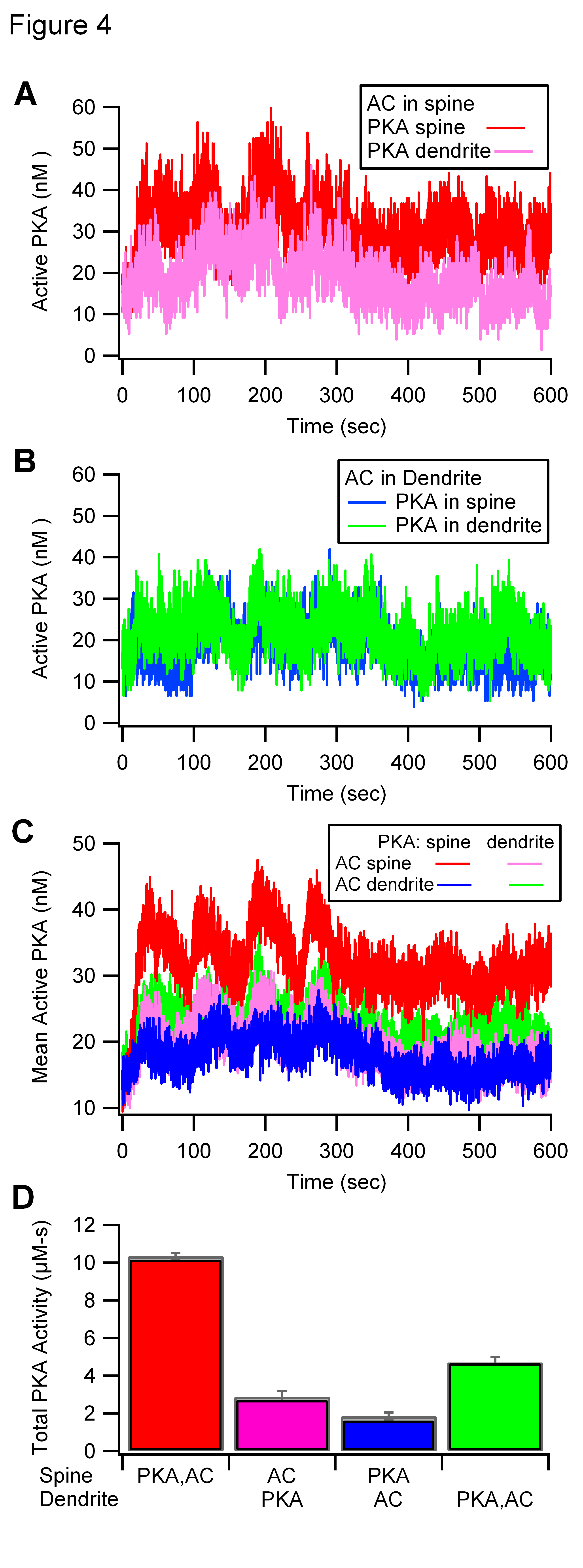 Figure 5 shows different molecules output from these and two
additional simulations (ModelA = spine/spine, ModelE = dend/dend,
Model B = spine/dend, Model D = dend/spine). Most other figures show
traces averaged over multiple random seeds. I hope that these model
use the random seed in these figures, but I don't guarantee it.
The README file for NeuroRD is also supplied under NeuroRD-README.txt
Figure 5 shows different molecules output from these and two
additional simulations (ModelA = spine/spine, ModelE = dend/dend,
Model B = spine/dend, Model D = dend/spine). Most other figures show
traces averaged over multiple random seeds. I hope that these model
use the random seed in these figures, but I don't guarantee it.
The README file for NeuroRD is also supplied under NeuroRD-README.txt
 Above Figure 3 shows the output from two simulations.
Above Figure 3 shows the output from two simulations.
 Figure 5 shows different molecules output from these and two
additional simulations (ModelA = spine/spine, ModelE = dend/dend,
Model B = spine/dend, Model D = dend/spine). Most other figures show
traces averaged over multiple random seeds. I hope that these model
use the random seed in these figures, but I don't guarantee it.
The README file for NeuroRD is also supplied under NeuroRD-README.txt
Figure 5 shows different molecules output from these and two
additional simulations (ModelA = spine/spine, ModelE = dend/dend,
Model B = spine/dend, Model D = dend/spine). Most other figures show
traces averaged over multiple random seeds. I hope that these model
use the random seed in these figures, but I don't guarantee it.
The README file for NeuroRD is also supplied under NeuroRD-README.txt