Files to reproduce the figures from:
Orio P, Soudry D (2012) Simple, fast and accurate implementation of
the diffusion approximation algorithm for stochastic ion channels with
multiple States PLoS One 7(5):e36670
Vclamp folder:
--------------
Scilab files to generate voltage clamp simulations with the K channel
from H&H model. Figures 2, 3 and 10A
- StochHH_K2 MC Vclamp.sci : MC modeling, uncoupled particles (2A)
- StochHH_K2 DA Vclamp.sci : DA algorithm, uncoupled particles (2B)
- StochHH_K2 DAss Vclamp.sci : DA with steady state app, uncoupled
particles (3A)
- StochHH_K2 Lss Vclamp.sci : Linaro et al. algorithm (10A)
- StochHH_K5 MC Vclamp.sci : MC modeling, coupled particles (2C)
- StochHH_K5 DA Vclamp.sci : DA algorithm, coupled particles (2D)
- StochHH_K5 DAGss Vclamp.sci : Goldwyn DA algorithm, s.s app (10A)
- StochHH_K5 DAss Vclamp.sci : DA with steady state app, coupled
particles (3B)
- StochHH_K5 DAG Vclamp.sci : Goldwyn DA algorithm, NO s.s app
Rb model folder:
----------------
Scilab/Matlab files to generate simulations of the Rubinstein's
mammalian Ranvier node model.
Figures 4-6 and 10B.
When run, each script will generate an output file with 4 columns:
Stimulus amplitude, Firing Efficiency, Mean Firing Time and FT
variance. The name of the file will indicate algorithm, number of
channels, dt, and real time elapsed.
- detRb 2vs8.sci: This will compare the deterministic version of the
model with coupled and uncoupled particles (no output).
- StochRb2 MC multi.sci: MC modeling, uncoupled particles
- StochRb2 DA multi.sci: DA algorithm, uncoupled particles
- StochRb2 DAss multi.sci: DA with steady state app, uncoupled
particles
- StochRb2 Lss multi.sci: Linaro et al. algorithm
- StochRb8 MC multi.sci: MC modeling, coupled particles
- StochRb8 DA multi.sci: DA algorithm, coupled particles
- StochRb8 DAGss multi.sci: Goldwyn DA algorithm, s.s. app.
- StochRb8 DAss multi.sci: DA with steady state app, coupled particles
- StochRb8 DAG multi.sci: Goldwyn DA algorithm, NO s.s. app.
HH model folder:
----------------
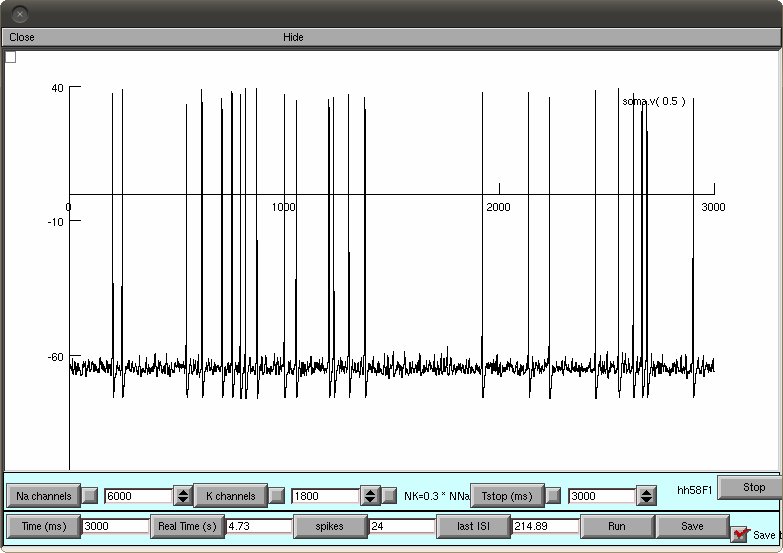
Neuron files to run the stochastic H&H models (figure 7-9). Compile
the supplied .mod files and run mosinit.hoc with nrngui. A prompt
window will appear to chose the simulation algorithm and the type of
simulation. If the default configuration of a DA model with coupled
particles is selected (click accept), and run is pressed, the
simulation should produce a graph similar to Figure 5 top right:
 Some Matlab files are also provided.
Some Matlab files are also provided.
 Some Matlab files are also provided.
Some Matlab files are also provided.
Some Matlab files are also provided.