This is a readme for the model associated with our paper:
Matsui H, Zheng M, Hoshino O (2014)
Facilitation of neuronal responses by intrinsic default mode network activity.
Neural Comput. 2014 Nov;26(11):2441-64.
This model simulates how default mode network activity affects sensory
information processing. The code was developed by Hiroakira Matsui.
The model is presented in C code and contains the following files:
*DMN.c :
The program source code.
*MT19937.h :
Header file of random number generator.
This is free file of Mersenne twister algorism.
(http://www.math.sci.hiroshima-u.ac.jp/~m-mat/MT/MT2002/emt19937ar.html)
*parameter.h :
All parameters to be required with this model are included.
Especially, you can regulate strength of tonic and phasic excitation
by changing of 'w_Ia_DMN' and 'w_p_DMN_Nsen' in this file,
respectively.
Usage:
After the putting all files in the same directory, compile DMN.c and
run it. Sample linux compile and run commands at the shell prompt are:
gcc DMN.c -Ilib -lm -o DMN
./DMN
This program makes following files.
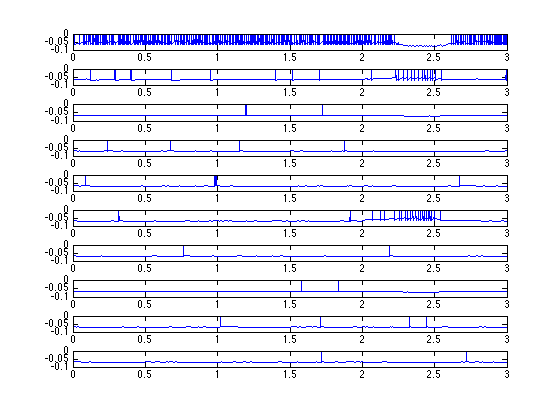
*data.csv :
This file shows membrane potentials of neurons and concentrations of
ambient-GABA. You can check the network and ambient-GABA transport
dynamics.
An example graph of the first 10 traces can be plotted in matlab with
a few lines: first on the linux command line:
$ wc data.csv
30001 210007 20280323 data.csv
$ tail -30000 data.csv > d.csv
Then on the matlab command prompt:
load d.csv
t=(0:length(d)-1)*1e-4;
figure
for i=1:10
subplot(10,1,i)
plot(t,d(:,i))
end
 *raster_plot.dat :
This file is raster plots of P cells. Please load this file into data
analysis software and plot. It shows how long the network takes time
to respond and which cell assembly responds to input. Repeating the
simulation in different onset input times, you can get reaction time
and error rate.
*up.csv :
This file shows membrane potentials of P cells of Nsen for
ongoing-spontaneous time period. Please load this file into
spreadsheet software and get membrane potential average and histogram.
*raster_plot.dat :
This file is raster plots of P cells. Please load this file into data
analysis software and plot. It shows how long the network takes time
to respond and which cell assembly responds to input. Repeating the
simulation in different onset input times, you can get reaction time
and error rate.
*up.csv :
This file shows membrane potentials of P cells of Nsen for
ongoing-spontaneous time period. Please load this file into
spreadsheet software and get membrane potential average and histogram.
 *raster_plot.dat :
This file is raster plots of P cells. Please load this file into data
analysis software and plot. It shows how long the network takes time
to respond and which cell assembly responds to input. Repeating the
simulation in different onset input times, you can get reaction time
and error rate.
*up.csv :
This file shows membrane potentials of P cells of Nsen for
ongoing-spontaneous time period. Please load this file into
spreadsheet software and get membrane potential average and histogram.
*raster_plot.dat :
This file is raster plots of P cells. Please load this file into data
analysis software and plot. It shows how long the network takes time
to respond and which cell assembly responds to input. Repeating the
simulation in different onset input times, you can get reaction time
and error rate.
*up.csv :
This file shows membrane potentials of P cells of Nsen for
ongoing-spontaneous time period. Please load this file into
spreadsheet software and get membrane potential average and histogram.