# ASTRO Fig 5 K⁺ Dynamics in Fully Reconstructed Four CA1 Astrocytes
## Description
This package implements the model described in *Savtchenko et al. (2018, Nature Communications)*, Figure 5, simulating extracellular K⁺ accumulation and clearance in a network of four fully reconstructed CA1 astrocytes using the ASTRO builder.
## Requirements
- **NEURON** v7.8 or later (with Python support)
- **Python** 3.7+
## Installation
1. Install NEURON with Python bindings (see [https://nrn.readthedocs.io](https://nrn.readthedocs.io)).
2. Clone or unzip this folder into your working directory.
3. From within the `mechanisms/` directory, compile the MOD files. With recent NEURON versions, you can do this by running `nrnivmodl MOD_files` from a terminal in the top-level directory.
> **Note:** Ensure that your `PYTHONPATH` or Python virtual environment includes NEURON’s `lib/python` directory.
## Usage
1. Open a terminal in the top-level directory.
2. Launch NEURON’s GUI with the initialization script:
```
nrngui init.hoc
```
3. You will see several windows/panels:
### Potassium Input Simulation
- Located on the left.
- Enter your extracellular K⁺ source parameters:
- **X, Y, Z** coordinates (µm)
- **Input radius** (µm)
- **Input onset** (ms) and **end** (ms)
- **[K⁺]out** (mM)
-
Click **Run simulation** once your sphere is positioned.
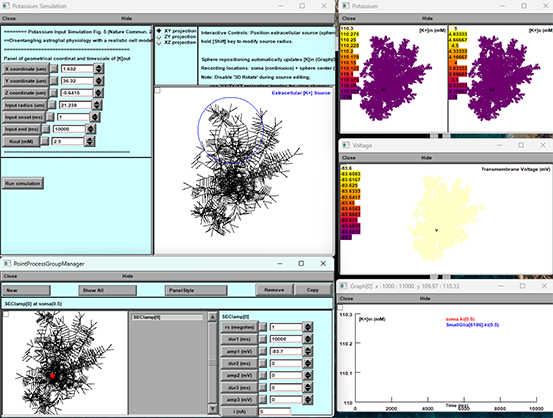
### 3D Morphology View
- Center panel.
- Displays reconstructed astrocyte morphology and the K⁺ source sphere.
### Potassium Panel
- Top-right.
- Two heatmaps:
- `[K⁺]in (mM)` inside the cell
- `[K⁺]o (mM)` outside the cell
- Color scale updates in real time.
### Voltage Panel
- Middle-right.
- Membrane voltage (mV) across the morphology.
### Graph[0] Panel
- Bottom-right.
- Time-series plot of selected variables (e.g., soma [K⁺]in vs. [K⁺]o).
### PointProcessGroupManager
- Bottom panel.
- Lists SEClamp or other point processes; control buttons available here.
4. Position your spherical K⁺ source by either:
- Typing new coordinates/radius in the **Potassium Input Simulation** panel, or
- Clicking and dragging the sphere in the **3D Morphology View** (hold **Shift** to resize).
5. Once your stimulus is set, press **Run simulation**.
6. Watch the **Potassium**, **Voltage**, and **Graph[0]** panels update to show how extracellular [K⁺] drives changes in membrane voltage and intracellular [K⁺].