This is the read me file for the dorsal striatum medium spiny
projection neuron from:
Evans, R.C., Morera-Herreras, T., Cui, Y., Du, K., Sheehan, T.,
Kotaleski, J.H., Venance, L., and Blackwell, K.T. (2012).
The Effects of NMDA Subunit Composition on Calcium Influx and Spike
Timing-Dependent Plasticity in Striatal Medium Spiny Neurons.
PLoS Comput. Biol. 8, e1002493.
Abstract:
Calcium through NMDA receptors (NMDARs) is necessary for the long-term
potentiation (LTP) of synaptic strength; however, NMDARs differ in
several properties that can influence the amount of calcium influx
into the spine. These properties, such as sensitivity to magnesium
block and conductance decay kinetics, change the receptor's response
to spike timing dependent plasticity (STDP) protocols, and thereby
shape synaptic integration and information processing. This study
investigates the role of GluN2 subunit differences on spine calcium
concentration during several STDP protocols in a model of a striatal
medium spiny projection neuron (MSPN). The multi-compartment,
multi-channel model exhibits firing frequency, spike width, and
latency to first spike similar to current clamp data from mouse dorsal
striatum MSPN. We find that NMDAR-mediated calcium is dependent on
GluN2 subunit type, action potential timing, duration of somatic
depolarization, and number of action potentials. Furthermore, the
model demonstrates that in MSPNs, GluN2A and GluN2B control which STDP
intervals allow for substantial calcium elevation in spines. The model
predicts that blocking GluN2B subunits would modulate the range of
intervals that cause long term potentiation. We confirmed this
prediction experimentally, demonstrating that blocking GluN2B in the
striatum, narrows the range of STDP intervals that cause long term
potentiation. This ability of the GluN2 subunit to modulate the shape
of the STDP curve could underlie the role that GluN2 subunits play in
learning and development.
Usage:
Open Genesis in a Linux environment (also works in Cygwin).
To run the model, type "MSsimspine" at Genesis prompt. This makes the
cell and a graph, but does not actually run the simulation.
To run a simulation type "include if2.g"; at end should see:
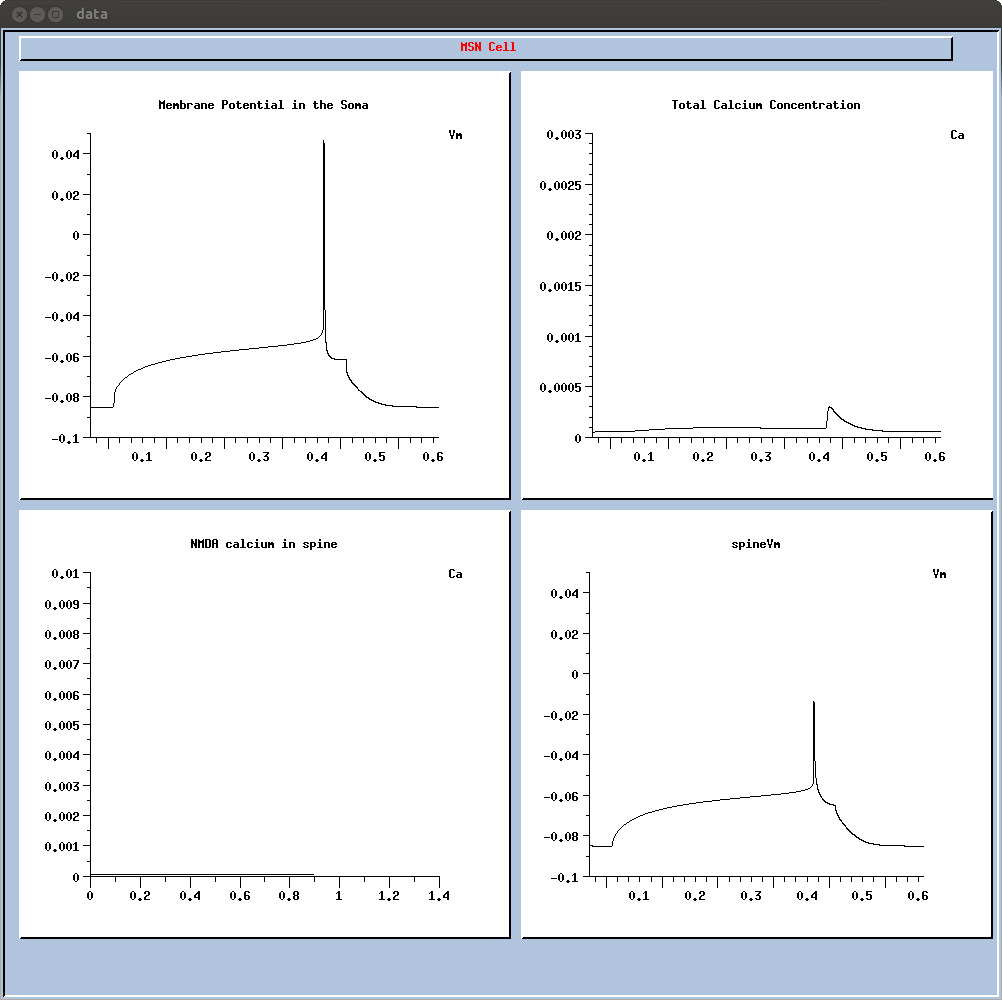 This will give a 400ms depolarization resulting in a single spike with
a long latency (figure 1B left). If you want to change the
depolarization level (to generate other curves from fig 1C left),
change the 'inj' parameter in if2.g.
To run the 30ms STDP protocols, make the cell and then type "include
30negsec.g"; "include 30posFillin.g"; or "include
30possec.g". Together these run the whole set of deltaTs; the green
traces in fig 2B are a subset of those produces by 30posFillin.g. The
NMDA calcium in one of the stimulated secondary dendritic spines
during these simulations was extracted to make figure 2C (green
trace). The deltaT (x axis in Figure 2C) was determined by manually
extracting the peak of the AP in each trace and comparing it to the
time of the stimulation.
To run simulations with a different NMDA subunit (as shown in Figure
3), open MScell/MScellspine.g, and edit the line "include
MScell/SynParamsCtx.g" to include SynParamsA.g (for NR2A),
SynParamsB.g (for NR2B) etc. If synparamsA are chosen, the NMDA receptor
will be "NR2A", while if Synparamsctx is chosen, the NMDA receptor will
be called "cortex". In addition, the if2.g simulation will not show any
NMDA calcium, be cause it contains no pre-synaptic activity.
This will give a 400ms depolarization resulting in a single spike with
a long latency (figure 1B left). If you want to change the
depolarization level (to generate other curves from fig 1C left),
change the 'inj' parameter in if2.g.
To run the 30ms STDP protocols, make the cell and then type "include
30negsec.g"; "include 30posFillin.g"; or "include
30possec.g". Together these run the whole set of deltaTs; the green
traces in fig 2B are a subset of those produces by 30posFillin.g. The
NMDA calcium in one of the stimulated secondary dendritic spines
during these simulations was extracted to make figure 2C (green
trace). The deltaT (x axis in Figure 2C) was determined by manually
extracting the peak of the AP in each trace and comparing it to the
time of the stimulation.
To run simulations with a different NMDA subunit (as shown in Figure
3), open MScell/MScellspine.g, and edit the line "include
MScell/SynParamsCtx.g" to include SynParamsA.g (for NR2A),
SynParamsB.g (for NR2B) etc. If synparamsA are chosen, the NMDA receptor
will be "NR2A", while if Synparamsctx is chosen, the NMDA receptor will
be called "cortex". In addition, the if2.g simulation will not show any
NMDA calcium, be cause it contains no pre-synaptic activity.
 This will give a 400ms depolarization resulting in a single spike with
a long latency (figure 1B left). If you want to change the
depolarization level (to generate other curves from fig 1C left),
change the 'inj' parameter in if2.g.
To run the 30ms STDP protocols, make the cell and then type "include
30negsec.g"; "include 30posFillin.g"; or "include
30possec.g". Together these run the whole set of deltaTs; the green
traces in fig 2B are a subset of those produces by 30posFillin.g. The
NMDA calcium in one of the stimulated secondary dendritic spines
during these simulations was extracted to make figure 2C (green
trace). The deltaT (x axis in Figure 2C) was determined by manually
extracting the peak of the AP in each trace and comparing it to the
time of the stimulation.
To run simulations with a different NMDA subunit (as shown in Figure
3), open MScell/MScellspine.g, and edit the line "include
MScell/SynParamsCtx.g" to include SynParamsA.g (for NR2A),
SynParamsB.g (for NR2B) etc. If synparamsA are chosen, the NMDA receptor
will be "NR2A", while if Synparamsctx is chosen, the NMDA receptor will
be called "cortex". In addition, the if2.g simulation will not show any
NMDA calcium, be cause it contains no pre-synaptic activity.
This will give a 400ms depolarization resulting in a single spike with
a long latency (figure 1B left). If you want to change the
depolarization level (to generate other curves from fig 1C left),
change the 'inj' parameter in if2.g.
To run the 30ms STDP protocols, make the cell and then type "include
30negsec.g"; "include 30posFillin.g"; or "include
30possec.g". Together these run the whole set of deltaTs; the green
traces in fig 2B are a subset of those produces by 30posFillin.g. The
NMDA calcium in one of the stimulated secondary dendritic spines
during these simulations was extracted to make figure 2C (green
trace). The deltaT (x axis in Figure 2C) was determined by manually
extracting the peak of the AP in each trace and comparing it to the
time of the stimulation.
To run simulations with a different NMDA subunit (as shown in Figure
3), open MScell/MScellspine.g, and edit the line "include
MScell/SynParamsCtx.g" to include SynParamsA.g (for NR2A),
SynParamsB.g (for NR2B) etc. If synparamsA are chosen, the NMDA receptor
will be "NR2A", while if Synparamsctx is chosen, the NMDA receptor will
be called "cortex". In addition, the if2.g simulation will not show any
NMDA calcium, be cause it contains no pre-synaptic activity.