This is the readme for the model associated with the paper
"Effects of Neural Morphology and Input Distribution on Synaptic Processing by Global and Focal NMDA-spikes" by Alon Poleg-Polsky. PLoS, accepted
The simulation examines the outcome of glutamatergic activation in different principal cortical cells (layer 2/3 pyramidal, layer 4 spiny stellate, layer 5b pyramidal). Clustered glutamatergic inputs were shown experimentally to trigger focal NMDA spikes in all of these cells types. This spike was hypothesized to mediate a multicompartmental computational processing, as activation of inputs in two branches was found to summate independently (Polsky et al 2004). However, randomly distributed glutamatergic drive can also elicit a different, global type of NMDA spike (Lavzin et al 2012). Here I examined the transition beyond the focal to global NMDA spike. How many synapses are needed to generate each spike in different cells? What is the local and somatic impact? How many branches need to be activated together before inter-branch interactions occur?
The simulation predicts that the morphology of a neuron can have a significant influence on the dendritic spike it can produce. Superficial layers neurons are more likely to generate global NMDA spikes, and synapses on neighboring branches interact early on. Layer 5 neurons have a significantly lower probability to generate a global NMDA spike and although synaptic interactions can occur between 3 and more branches, the number of action potentials follows the linear summation prediction.
Running the simulation:
First select the neuron to simulate from MAIN.hoc, line 2. The simulation has the 5 layer2/3, 5 layer 4 and 4 layer 5(V) cells. In pyramidal cell morphologies basal and apical dendrites are separated by their name; dend[x] are basal, apic[x] are apical dendrites. The user needs to add the dendritic tree to the dends SectionList. Example of inclusion of the basal tree would be:
for i=0,69{dend[i] dends.append()}
An apical tree:
access soma
for i=0,115{
access
apic[i]
if
(distance(1)>500){ dends.append()}
}
To include spiking, please set the 'insertHH' variable to 1.
GUI:
'Parameters' panel:
#MAX-the number of glutamatergic synapses in a simulation. When this filed is changed by the user distributes the synapses randomly over the selected branches.
NMDA,AMPA(nS): the unitary synaptic conductance
Active Params: when spiking is included sets the voltage gated channel conductances at the soma and dendrites
'Between' panel:
In each cell, the simulation finds all the terminal dendrites. The user can set the number of synapses activated on each one. To test synaptic interactions between branches then, first set a desired number of synaptic inputs on each of the examined dendrites individually, run the simulation and record the responses. Afterwards activate synapses on all of the selected branches and compare the resulting response to the predicted summation of individual branches.
Below are examples from a layer 2/3 simulation (layer23_5 cell)
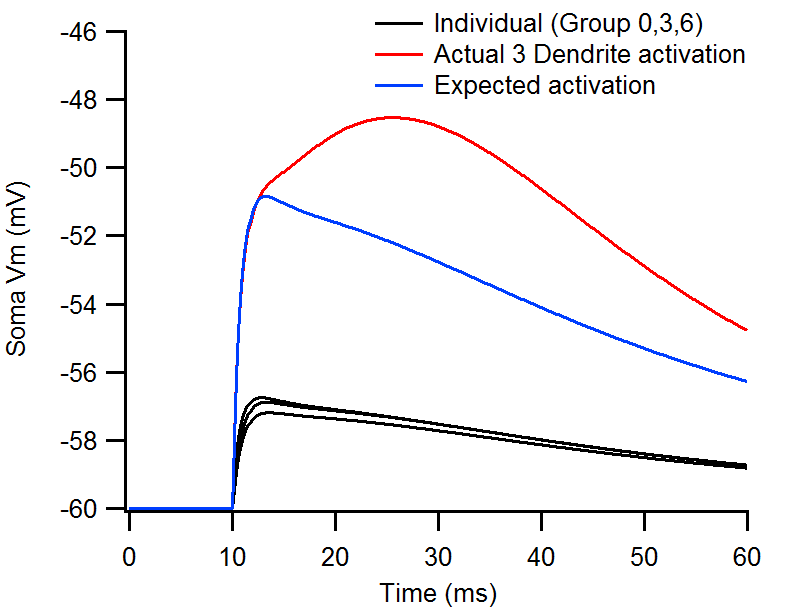
Activation of 25 synapses on each one of three basal dendrites (groups 0, 3, 6) results in 3-4mV somatic EPSPs (black). When activated together, the EPSP reaches 12mV (red), well above the predicted linear summation of the three branch EPSPs (blue).

Somatic EPSPs for randomly distributed synaptic input from the same cell demonstrating a global NMDA spike. red NMDARs + AMPARs, black, AMPARs only. The number of activated synapses ranged from 10 to 190 in steps of 20.

Comparison of sub- and supra-threshold responses. Red global NMDA spike (150 randomly distributed synapses) in a passive cell. Yellow, the same spike with voltage gated potassium channels. Blue, same synaptic activation with somatic voltage gated sodium channels.