This simulation was tested/developed on LINUX systems, but may run on Microsoft Windows or Mac OS. To run, you will need the NEURON simulator (available at http://www.neuron.yale.edu) compiled with python enabled. Unzip the contents of the zip file to a new directory. compile the mod files from the command line with: nrnivmodl *.mod That will produce an architecture-dependent folder with a script called special. On 64 bit systems the folder is x86_64. To run the simulation from the command line: ./x86_64/special -python mosinit.py then NEURON will start with the python interpreter and load the mechanisms and simulation. Next, the network and inputs will be setup. Then the simulation will run 40,000 times for 10,000 random initializations of network state and 4 input conditions (all combinations of TNF and GF ON/OFF). Once the simulation has run to completion (a few minutes on an Intel Xeon 2.27 GHz CPU), the output will be displayed in textual format. The output measures how often the network reaches an apoptotic state (apoptosis ratio), in each of the 4 input conditions, under the random initializations to network state. In this model, GF represents a growth factor (pro-survival) and TNF represents tumor necrosis factor (pro-apoptotic). The output will have values similar to these ([1]): TNF: 0 GF: 0 apop ratio: 0.4733 TNF: 0 GF: 1 apop ratio: 0.4679 TNF: 1 GF: 0 apop ratio: 0.9715 TNF: 1 GF: 1 apop ratio: 0.6398 Visualization of network state: The software provided makes use of the Graphviz and ImageMagick software packages (if they are already installed) to draw the network state over time, as an animated gif. An example of how to do this in a simulation loop is in the myanim function in network.py . myanim also requires a directory to store the individual frames in. This directory should be called frames, and be present in the current working directory. A temporary file, __junk__.dot will be written to as well. An example rendering of a network and its state is provided here:
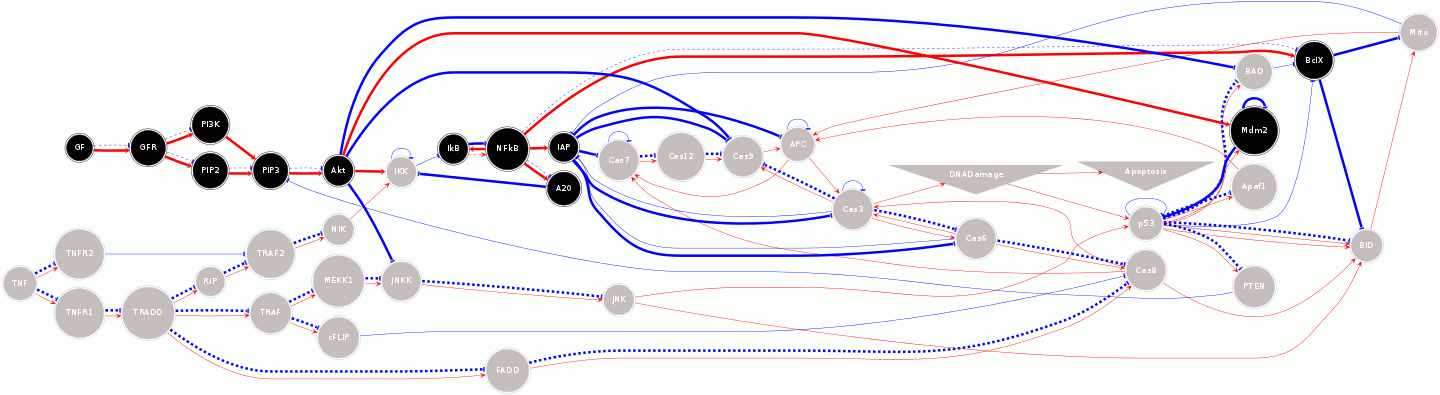 In this rendering, regular (special) nodes are drawn with circles
(triangles) with the name of the node indicated. Color represents node
state (off:gray, on:black). Solid (dotted) lines represent that a
source node must be ON (OFF) to activate the rule. Lines indicate
interaction rules (red:activating; blue:inhibiting). Thick (thin)
lines indicate a currently active (inactive) rule.
References:
This simulation is based on the article:
[1] Boolean network-based analysis of the apoptosis network:
Irreversible apoptosis and stable surviving Journal of Theoretical
Biology volume 259, pages 760-769. by Mai Z and Liu, H (2009).
For questions/comments email: samuel dot neymotin at yale dot edu or
samn at neurosim dot downstate dot edu
Changelog:
20160920 This updated version from the Lytton lab allows this model to
run on the mac.
20220523 Updated MOD files to contain valid C++ and be compatible with
the upcoming versions 8.2 and 9.0 of NEURON. Updated to use post ~2011
signature of mcell_ran4_init function.
20221216 Python3 migration via 2to3.
In this rendering, regular (special) nodes are drawn with circles
(triangles) with the name of the node indicated. Color represents node
state (off:gray, on:black). Solid (dotted) lines represent that a
source node must be ON (OFF) to activate the rule. Lines indicate
interaction rules (red:activating; blue:inhibiting). Thick (thin)
lines indicate a currently active (inactive) rule.
References:
This simulation is based on the article:
[1] Boolean network-based analysis of the apoptosis network:
Irreversible apoptosis and stable surviving Journal of Theoretical
Biology volume 259, pages 760-769. by Mai Z and Liu, H (2009).
For questions/comments email: samuel dot neymotin at yale dot edu or
samn at neurosim dot downstate dot edu
Changelog:
20160920 This updated version from the Lytton lab allows this model to
run on the mac.
20220523 Updated MOD files to contain valid C++ and be compatible with
the upcoming versions 8.2 and 9.0 of NEURON. Updated to use post ~2011
signature of mcell_ran4_init function.
20221216 Python3 migration via 2to3.