This is the corrected version. CaBK.mod and na.mod have been modified
as per the description in forum (see link below).
Both the original and the corrected versions are available here in
ModelDB. Note that the cell models in the corrected version have not
yet been tuned to take into account the behavior of the corrected ion
channel.
Note: please see the following NEURON forum post describing how bugs
found in a few of the ion channel models were fixed:
https://www.neuron.yale.edu/phpBB/viewtopic.php?f=18&t=2956
This model was implemented by Rob Morgan in the Soltesz lab at UC
Irvine. It is a scaleable dentate gyrus model including four cell
types. This model runs in serial (on a single processor) and has been
published at the size of 50,000 granule cells (with proportional
numbers of the other cells).
********************
The relevant publications are:
Morgan, R.J. and I. Soltesz, Nonrandom connectivity of the epileptic
dentate gyrus predicts a major role for neuronal hubs in
seizures. Proc Natl Acad Sci U S A, 2008. 105(16): p. 6179-84.
Dyhrfjeld-Johnsen, J., et al., Topological determinants of
epileptogenesis in large-scale structural and functional models of the
dentate gyrus derived from experimental data. J Neurophysiol,
2007. 97(2): p. 1566-87.
Morgan, R.J., V. Santhakumar, and I. Soltesz, Modeling the dentate
gyrus. Prog Brain Res, 2007. 163: p. 639-58.
Santhakumar, V., I. Aradi, and I. Soltesz, Role of mossy fiber
sprouting and mossy cell loss in hyperexcitability: a network model of
the dentate gyrus incorporating cell types and axonal topography. J
Neurophysiol, 2005. 93(1): p. 437-53.
********************
Directory contents:
*.mod: Mechanism files for ionic conductances
*cell.*cell: Synaptic parameters for connections between
precell.postcell
p*c.hoc: Connection probabilities for each cell type to the other cell
types in the network. Precell type is given by the middle letter in
the filename.
*cdist.hoc: Axonal arbor distributions used in determining connection
probabilities
50knet.hoc: Main neuron code for the network
run50knet.bash: Bash script for running the network (unix/linux).
This script needs to be modified to contain the actual paths for where
you placed the files.
********************
Running the Network
1) The mod files must first be compiled into the directory from which
you will run the network.
ex:
nrnivmodl ccanl nca tca LcaMig CaBK gskch hyperde3 bgka ichan2 Gfluct2
2) Run the file run50knet.bash. This file does several things:
a) Creates a directory according to the number of granule cells in the
network
b) Creates a parameters.dat file in the working directory which
contains such things as the number of cells for each cell type and the
percent of network sclerosis (the network is scalable to some degree
but if you alter the number of cells, you may also have to change the
connection probabilities and synaptic weights to ensure a
well-connected network with reasonable synaptic connectivities).
c) Runs the hoc code
d) Moves the two output files, numCons.dat and spikerast.dat to the
directory that was created in (a)
** While the hoc code for running the network includes usage of the
gui, it's highly recommended to run the program in the background
with no graphical display for large networks.
When run in its default mode (auto-launched from ModelDB for example)
the simulation will produce (about 2 minutes after the start button is
pressed) a figure like:
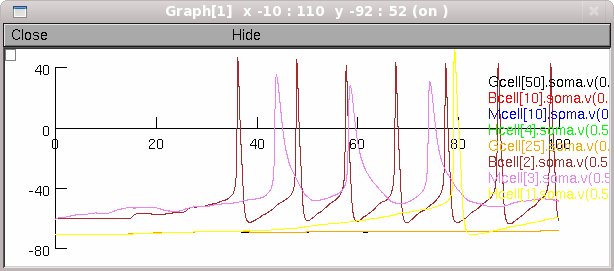 ---
Changelog
=========
* 20140311 Marianne Bezaire: added frecord_init() on line 1554 of
50knet.hoc so that vector recordings from cells will work properly in
case anyone wants to do that.
* 20220924: Update MOD files to avoid declaring variables and functions
with the same name. See https://github.com/neuronsimulator/nrn/pull/1992
* 20221124: hyperde3 and ichan2: drop INITIAL return values, just return
---
Changelog
=========
* 20140311 Marianne Bezaire: added frecord_init() on line 1554 of
50knet.hoc so that vector recordings from cells will work properly in
case anyone wants to do that.
* 20220924: Update MOD files to avoid declaring variables and functions
with the same name. See https://github.com/neuronsimulator/nrn/pull/1992
* 20221124: hyperde3 and ichan2: drop INITIAL return values, just return
 ---
Changelog
=========
* 20140311 Marianne Bezaire: added frecord_init() on line 1554 of
50knet.hoc so that vector recordings from cells will work properly in
case anyone wants to do that.
* 20220924: Update MOD files to avoid declaring variables and functions
with the same name. See https://github.com/neuronsimulator/nrn/pull/1992
* 20221124: hyperde3 and ichan2: drop INITIAL return values, just return
---
Changelog
=========
* 20140311 Marianne Bezaire: added frecord_init() on line 1554 of
50knet.hoc so that vector recordings from cells will work properly in
case anyone wants to do that.
* 20220924: Update MOD files to avoid declaring variables and functions
with the same name. See https://github.com/neuronsimulator/nrn/pull/1992
* 20221124: hyperde3 and ichan2: drop INITIAL return values, just return