FlyBrainLab is an interactive computing platform for studying the function of executable circuits constructed from fruit fly brain datasets.
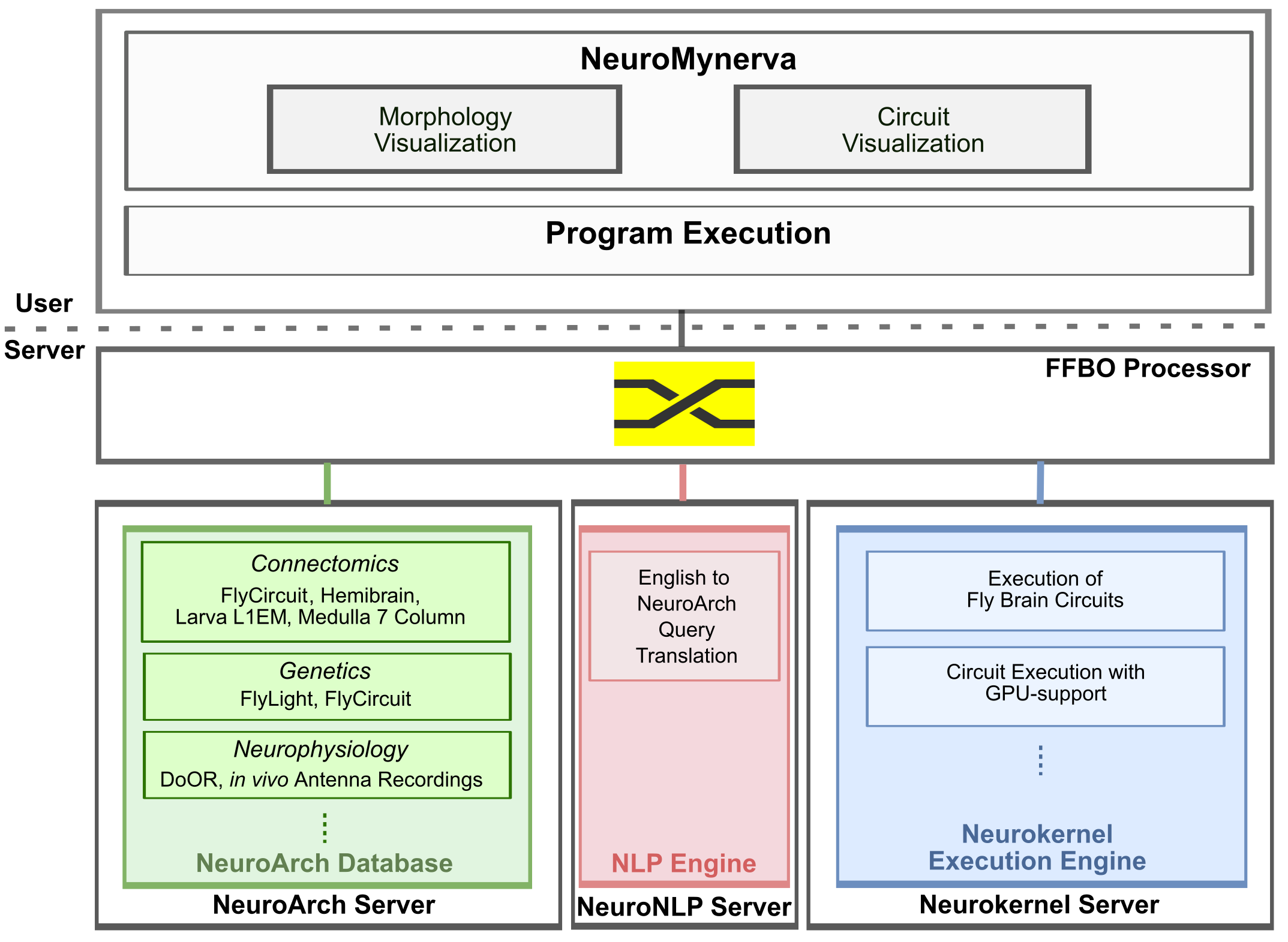
General information about the FlyBrainLab is available at https://www.fruitflybrain.org. Stable and tested FlyBrainLab installation instructions for Linux, MacOS, and Windows are available at https://github.com/FlyBrainLab/FlyBrainLab. A full installation of FlyBrainLab, including all backend and frontend components, is available as a Docker image at https://hub.docker.com/r/fruitflybrain/fbl. The image requires a Linux host with at least 1 CUDA-enabled GPU and the nvidia-docker package installed. For a custom installation of the complete FlyBrainLab platform, a shell script is available at https://github.com/FlyBrainLab/FlyBrainLab.The main components of the FlyBrainLab software architecture are depicted in the following figure.

The server-side components include the FFBO Processor, the NeuroArch Server, the NeuroNLP Server and the Neurokernel Server.
FFBO Processor implements a Crossbar.io router that establishes the communication path among connected components and clients. Components communicate using routed Remote Procedure Calls (RPCs) and a publish/subscribe mechanism. The FFBO processor can be hosted locally or in the cloud.
NeuroArch Server hosts the NeuroArch Database implemented with the OrientDB graph database. The NeuroArch Database provides a novel data model for representation and storage of connectomic, synaptomic, genetic and neurophysiology data of the fruit fly brain with cross-referenced executable circuits. The Queries of the NeuroArch Database are supported by the NeuroArch Python API. The NeuroArch Server provides high level RPC APIs for remote access of the NeuroArch Database.
Neurokernel Server provides RPC APIs for code execution of model circuits by the Neurokernel Execution Engine. Neurokernel supports the easy combination of independently developed executable circuits towards the realization of a complete whole brain emulation. The Neurokernel Execution Engine features:
The Neurokernel Server directly fetches the specification of executable circuits from the NeuroArch Server, instantiates these circuits and transfers them for execution to the Neurokernel Execution Engine.
NeuroNLP Server provides an RPC API for translating queries written as English sentences, such as "add dopaminergic neurons innervating the mushroom body", into database queries that can be interpreted by the NeuroArch Server API, using the NLP Engine hosted by NeuroArch_NLP library. This capability increases the accessibility of the NeuroArch Database to users without prior exposure to database programming, and greatly simplifies the often-demanding task of writing database queries.
| Repository | Utility | Source Code | Documentation |
|---|---|---|---|
| FFBO Processor | Main Processor and Router | GitHub | |
| NeuroArch Server | Database Server | GitHub | |
| NeuroArch | Graph Database | GitHub | Doc |
| NeuroNLP Server | Natural Language Processing Server | GitHub | |
| NeuroArch_NLP | Translation of English Query to NeuroArch Query | GitHub | |
| Neurokernel Server | Execution Server | GitHub | |
| Neurokernel | Execution Engine | GitHub | Doc |
| Neurodriver | Execution Engine | GitHub |
We’ve included a few publicly available datasets in the NeuroArch database for easy accessibility. Users can also load and use custom datasets with their local NeuroArch installations.
v1.2): Referencev1.0.1, v1.1, v1.2): ReferenceAdditionally, we provide a Datasets Version Tracker which includes information about and download links for the NeuroArch Database.
The FlyBrainLab user-side components consist of the NeuroMynerva User Interface and the FlyBrainLab Client back-end. The former provides capabilities such as morphology visualization and interactive circuit diagrams, and the latter provides program execution for communicating with the server components.
A more detailed software architecture of the user-side components is depicted below.
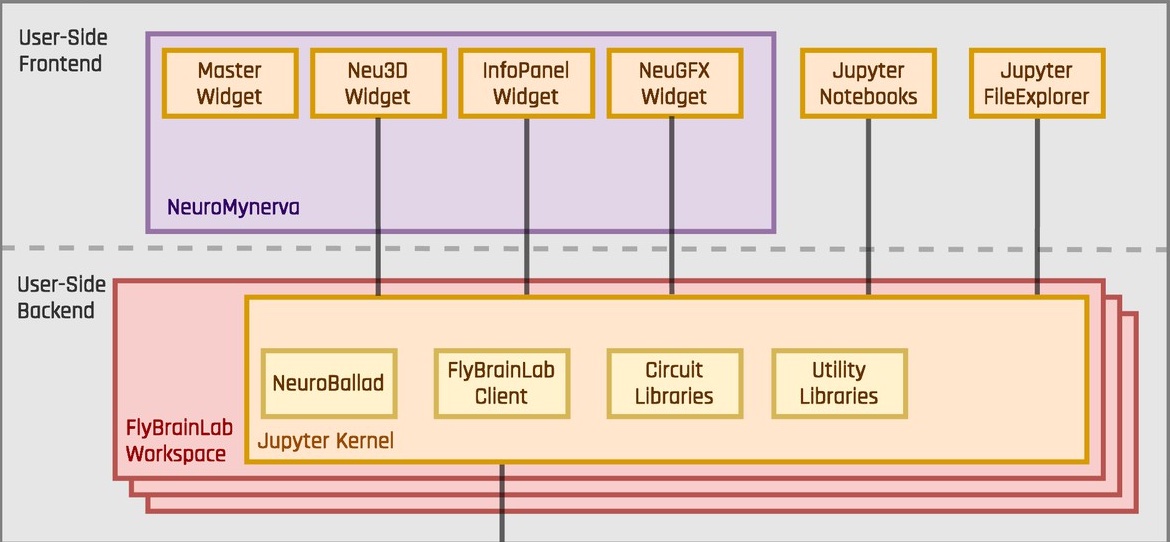
NeuroMynerva is the user-side frontend of FlyBrainLab. It is a browser-based application that substantially extends upon JupyterLab by providing a number of widgets, including a Neu3D widget for 3D visualization of fly brain data, a NeuGFX widget for exploring executable neural circuits with interactive circuit diagrams, and an InfoPanel widget for accessing individual neuron/synapse data.
FlyBrainLab Client back-end (FBLClient) connects to the FFBO processor and accesses services provided by the connected backend servers. FlyBrainLab Client provides program execution APIs for handling requests to the server-side components and parsing of data coming from backend servers. In addition, it handles the communication with the frontend through the Jupyter kernel.
Neuroballad is a Python library that simplifies and accelerates executable circuit construction and simulation using Neurokernel in Jupyter notebooks in FlyBrainLab.
FlyBrainLab Utility Library offers a number of utilities to explicate the graph structure of neural circuits from raw connectome and synaptome data.
FlyBrainLab Circuits Library offer circuit specific tools for analyzing, evaluating and comparing fruit fly brain circuits. In conjunction with the capability of visually constructing circuits enabled by the NeuroMynerva front-end, speeding up the process of creating interactive executable circuit diagrams can substantially reduce the exploratory development cycle.
FlyBrainLab User Components
| Repository | Utility | Source Code | Documentation |
|---|---|---|---|
| NeuroMynerva | User Frontend | GitHub | Doc |
| FBLClient | User Backend and FlyBrainLab Utility Library | GitHub | Doc |
| Neuroballad | High Level API for Circuit Execution | GitHub |
FlyBrainLab Utility Library
The current FlyBrainLab Utility Library includes:
They are built into the FBLClient.
FlyBrainLab Circuit Library
| Repository | Utility | Source Code | Documentation |
|---|---|---|---|
| CXcircuits | Library for the Central Complex | GitHub | |
| EOScircuits | Library for the Early Olfactory System | GitHub | Doc |
| OlfTrans | Library for Molecular Transduction in the Olfactory Sensory Neurons | GitHub | Doc |
| VisTrans | Library for Phototransduction in the Retina | GitHub |
Latest tutorials can be found in the FlyBrainLab/Tutorials repository.