The modFoss package for ion channel analysis as described in the paper
Ferneyhough GB, Thibealut CM, Dascalu SM, Harris FC (2016)
ModFossa: A library for modeling ion channels using Python.
J Bioinform Comput Biol 14:1642003
is available in github:
https://github.com/gareth-ferneyhough/modfossa
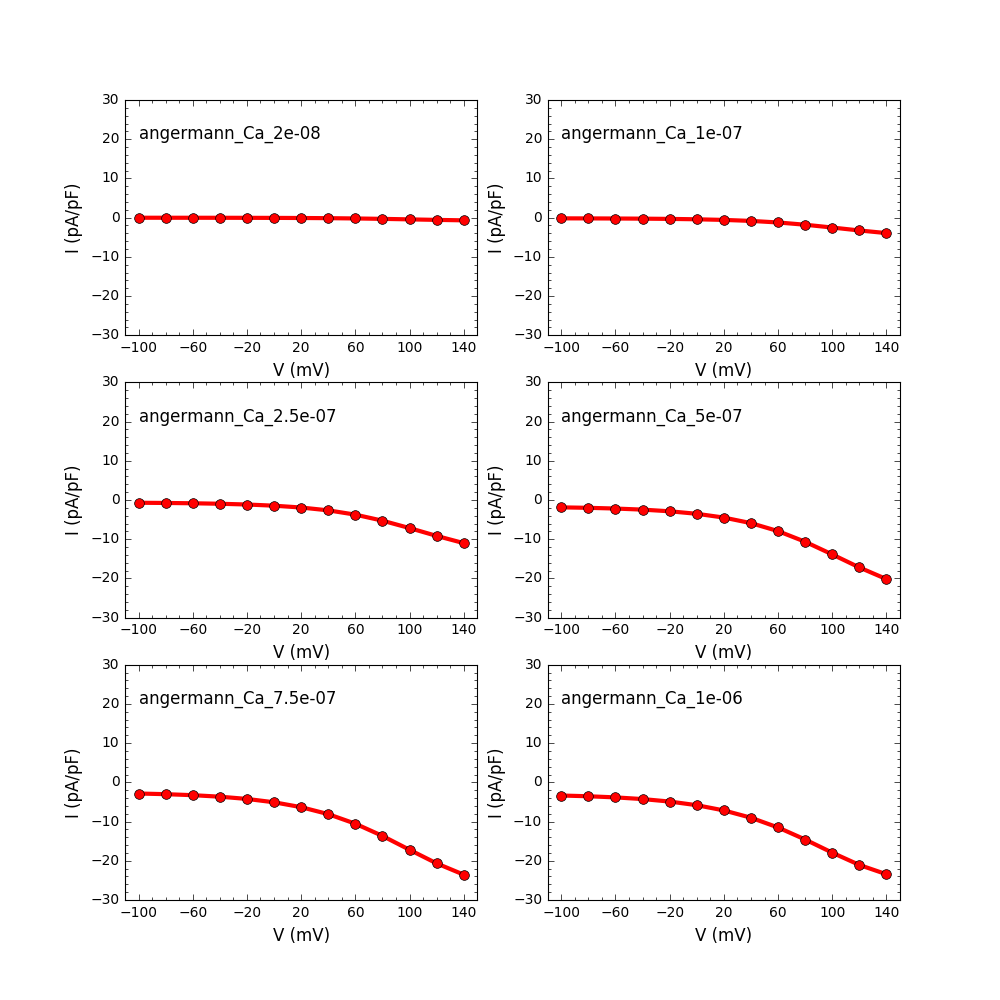
After building the python package you can run the examples provided in
the samples folder. For example running
python Angerman2006.py
and will produce graphs like:
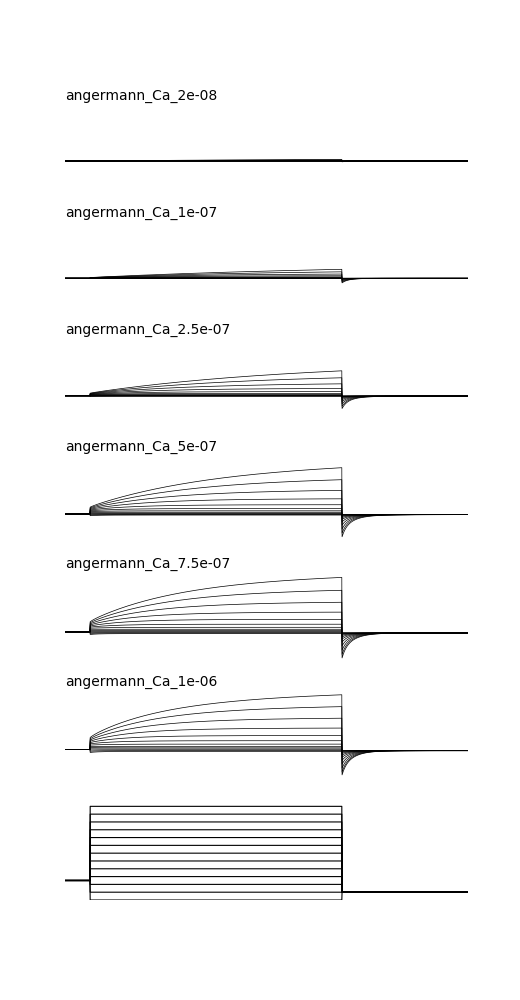
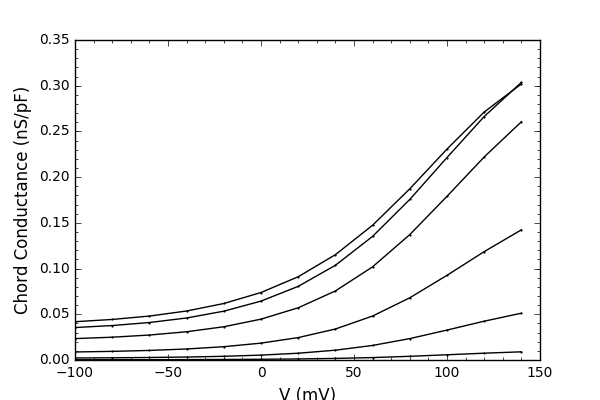
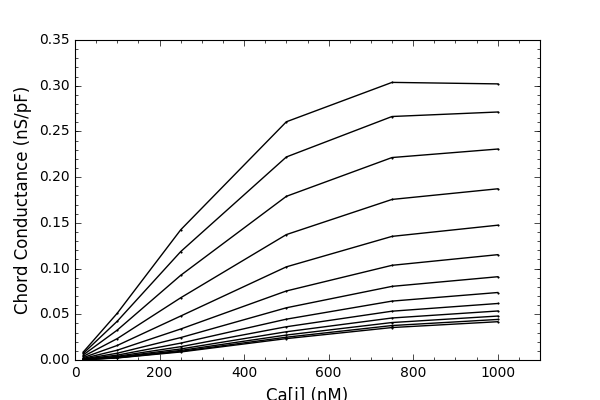
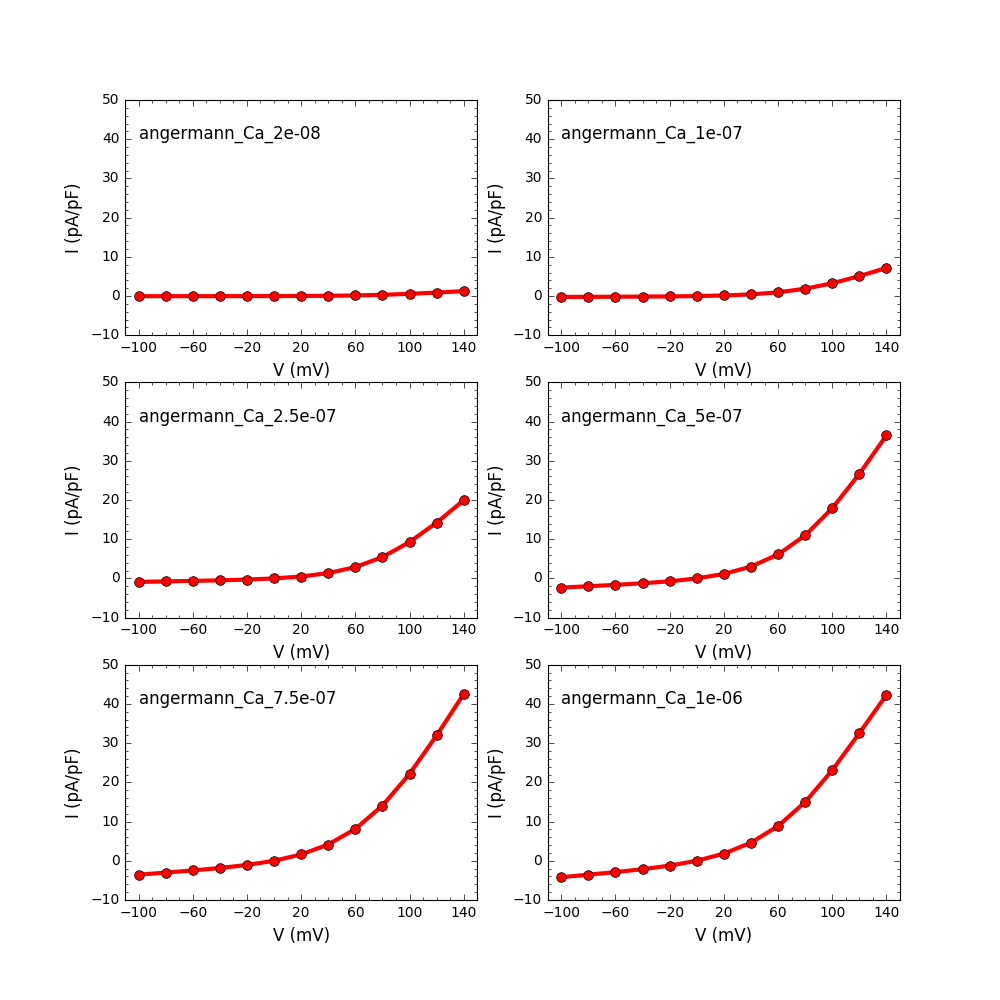
 Note from the ModelDB administrator:
When I was following the install instructions for modFoss on an ubuntu
16.04.01 in my home bin folder (/home/morse/bin) I found it helpful to
export these
export CPATH=/home/morse/bin/sundials-2.6.2/instdir/include/
export LIBRARY_PATH=/home/morse/bin/sundials-2.6.2/instdir/lib
in addition to their suggested
export LD_LIBRARY_PATH=/home/morse/bin/sundials-2.6.2/instdir/lib
Note from the ModelDB administrator:
When I was following the install instructions for modFoss on an ubuntu
16.04.01 in my home bin folder (/home/morse/bin) I found it helpful to
export these
export CPATH=/home/morse/bin/sundials-2.6.2/instdir/include/
export LIBRARY_PATH=/home/morse/bin/sundials-2.6.2/instdir/lib
in addition to their suggested
export LD_LIBRARY_PATH=/home/morse/bin/sundials-2.6.2/instdir/lib




 Note from the ModelDB administrator:
When I was following the install instructions for modFoss on an ubuntu
16.04.01 in my home bin folder (/home/morse/bin) I found it helpful to
export these
export CPATH=/home/morse/bin/sundials-2.6.2/instdir/include/
export LIBRARY_PATH=/home/morse/bin/sundials-2.6.2/instdir/lib
in addition to their suggested
export LD_LIBRARY_PATH=/home/morse/bin/sundials-2.6.2/instdir/lib
Note from the ModelDB administrator:
When I was following the install instructions for modFoss on an ubuntu
16.04.01 in my home bin folder (/home/morse/bin) I found it helpful to
export these
export CPATH=/home/morse/bin/sundials-2.6.2/instdir/include/
export LIBRARY_PATH=/home/morse/bin/sundials-2.6.2/instdir/lib
in addition to their suggested
export LD_LIBRARY_PATH=/home/morse/bin/sundials-2.6.2/instdir/lib