This is the readme for the models associated with the paper:
Sanjay M, Neymotin SA, Krothapalli SB (2015) Impaired dendritic
inhibition leads to epileptic activity in a computer model of
CA3. Hippocampus 25:1336-50
This simulation was tested/developed on LINUX systems, but may run on
Microsoft Windows or Mac OS.
To run, you will need the NEURON simulator (available at
http://www.neuron.yale.edu) compiled with python enabled.
The code ran with NEURON v7.2. Some problems have been noted with
NEURON v7.4 and fixes are under development to allow the code to run
with this newer version of NEURON.
Unzip the contents of the zip file to a new directory.
compile the mod files from the command line with:
nrnivmodl *.mod
That will produce an architecture-dependent folder with a script
called special. On 64 bit systems the folder is x86_64. To run the
simulation from the command line:
./x86_64/special -python mosinit.py
then NEURON will start with the python interpreter and load the
mechanisms and simulation. Next, the network and inputs will be setup.
Then the simulation will be run for 5 seconds of simulation time. The
simulation duration is modifiable via the h.tstop parameter in
params.py. Note that setup of the network may take 10-30 seconds,
depending on your processor speed and amount of RAM. Once the
simulation has run, two graphs will be displayed, showing local field
potential and individual cell responses and raster plot.
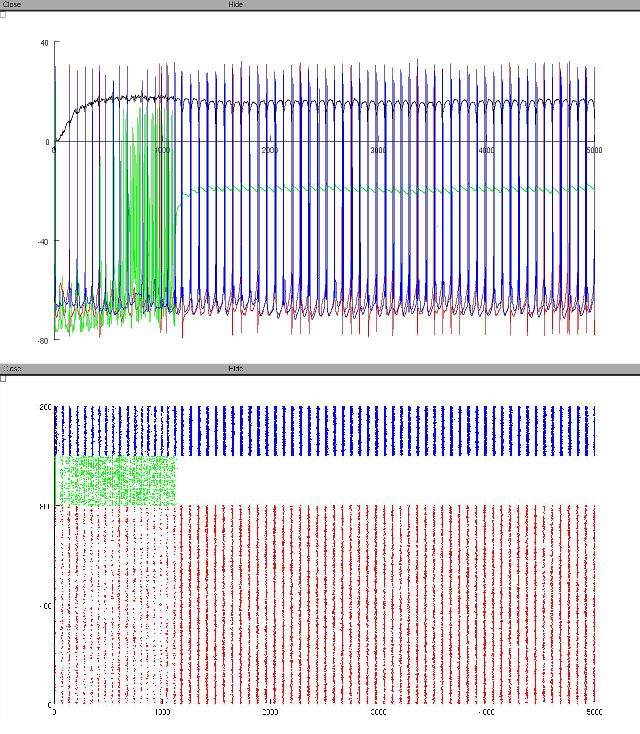 The spike raster is arranged with y-axis as cell identifier and x-axis
as time in milliseconds. The y-axis is further arranged in order of
cell-types (red=pyramidal, green=basket, blue=OLM).
For questions email: msanjaycmc at gmail dot com.
Changelog
---------
2022-05: Updated MOD files to contain valid C++ and be compatible with
the upcoming versions 8.2 and 9.0 of NEURON.
2022-12: Python3 migration via 2to3
The spike raster is arranged with y-axis as cell identifier and x-axis
as time in milliseconds. The y-axis is further arranged in order of
cell-types (red=pyramidal, green=basket, blue=OLM).
For questions email: msanjaycmc at gmail dot com.
Changelog
---------
2022-05: Updated MOD files to contain valid C++ and be compatible with
the upcoming versions 8.2 and 9.0 of NEURON.
2022-12: Python3 migration via 2to3
 The spike raster is arranged with y-axis as cell identifier and x-axis
as time in milliseconds. The y-axis is further arranged in order of
cell-types (red=pyramidal, green=basket, blue=OLM).
For questions email: msanjaycmc at gmail dot com.
Changelog
---------
2022-05: Updated MOD files to contain valid C++ and be compatible with
the upcoming versions 8.2 and 9.0 of NEURON.
2022-12: Python3 migration via 2to3
The spike raster is arranged with y-axis as cell identifier and x-axis
as time in milliseconds. The y-axis is further arranged in order of
cell-types (red=pyramidal, green=basket, blue=OLM).
For questions email: msanjaycmc at gmail dot com.
Changelog
---------
2022-05: Updated MOD files to contain valid C++ and be compatible with
the upcoming versions 8.2 and 9.0 of NEURON.
2022-12: Python3 migration via 2to3