Model of
multiplication in direction selective ganglion cells
NMDA Receptors Multiplicatively Scale Visual Signals and Enhance Directional Motion Discrimination in Retinal Ganglion Cells. Poleg-Polsky A, Diamond JS. Neuron. 2016 Mar 16;89(6):1277-90. doi: 10.1016/j.neuron.2016.02.013. Epub 2016 Mar 3. PMID:26948896
The simulation models synaptic integration in a multi-compartmental direction selective ganglion cell (DSGC).
The simulated DSGC receives 3 types of synaptic inputs:
(1) Glutamatergic drive, mediated by AMPA and NMDA receptors
(2) Cholinergic drive, mediated by nicotinic receptors
(3) GABAergic drive, mediated by GABAA receptors
The drives are activated in a spatial sequence, to imitate a moving stimulus. Preferred direction (PD) and null direction (ND) are different in the intensity of the synaptic drives. For tuned inhibition, the inhibitory drive is 300% larger in the ND. For tuned excitation, the cholinergic drive is larger by 300% in the PD. In all simulations the number of active glutamatergic synapses is similar between directions.
Three conditions are explored:
1. Control simulation, with tuned inhibition and voltage dependent NMDAR behavior
2. Zero Mg++ simulation, with tuned inhibition and voltage insensitive NDMARs
3. Tuned excitation simulation, with voltage dependent NMDAR behavior
In each condition clicking on the corresponding button with run either the PD or ND simulations.
NMDARs and voltage gated channels can be 'blocked' by unchecking the corresponding check boxes.
Figure 1: Control condition, PD activation w/ (black) an w/o (gray) NMDARs (spikes blocked)

Figure 2: Control condition, ND activation w/ (black) an w/o (gray) NMDARs (spikes blocked)

In the control case NMDARs amplify the response by about 100%, indicative of multiplication.
Voltage independent NMDARs amplify both PD (Figure 3) and ND (Figure 4) responses. The contribution of NMDARs becomes similar between PD and ND, indicative of additive scaling.
Figure 3: Voltage independent NMDARs, PD activation w/ (black) an w/o (gray) NMDARs (spikes blocked)

Figure 4: Voltage independent NMDARs, ND activation w/ (black) an w/o (gray) NMDARs (spikes blocked)

Similar additive scaling is observed with tuned excitation:
Figure 5: Tuned excitation, PD activation w/ (black) an w/o (gray) native NMDARs (spikes blocked)
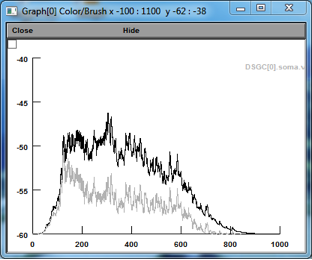
Figure 6: Tuned excitation, ND activation w/ (black) an w/o (gray) native NMDARs (spikes blocked)
