This multicompartment model of the cerebellar granule cell is
described in
Dover K, Marra C, Solinas S, Popovic M, Subramaniyam S, Zecevic D,
D'Angelo E, Goldfarb M (2016) FHF-independent conduction of action
potentials along the leak-resistant cerebellar granule cell axon.
Nat Commun 7:12895
and is a substantial revision of the prior model by Diwakar et al.,
Journal of Neurophysiology Published 1 February 2009 Vol. 101 no. 2,
519-532 DOI: 10.1152/jn.90382.2008
Link to prior model: (http://modeldb.yale.edu/116835).
The principal revisions are based upon genetic, biochemical, and axon
optical imaging and include: (1) very low leak conductance on the
distal axon and parallel fibers, and (2) differential tuning of the
voltage gated sodium conductance to reflect FHF modulation of gNav in
the hillock and axon initial segment, but not along the distal axon or
parallel fibers. When downloaded and opened, the model is preset to
simulate a 9 pA somatic current injection following a 60 msec delay,
which is necessarily incorporated to allow the state distribution in
the embedded Nav Markov model in each cellular compartment.
Paralleling the data in Figure 5 of Dover et al, the top window
corresponds to somatic voltage as in Fig. 5b,
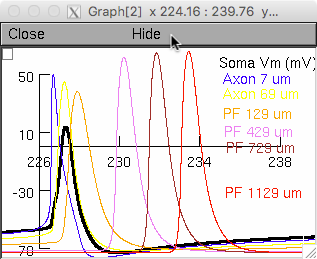
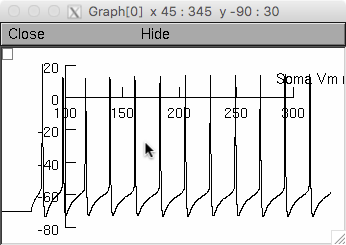 the middle tier windows correspond to traces showing the first and the
ninth induced spike conducting along the axon and parallel fiber as in
Fig 5d,
the middle tier windows correspond to traces showing the first and the
ninth induced spike conducting along the axon and parallel fiber as in
Fig 5d,
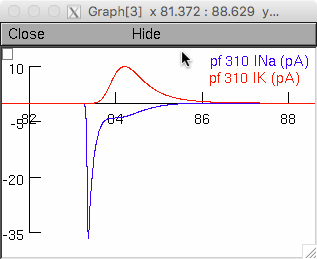
 and the bottom window shows sodium and potassium currents in a
parallel fiber compartment during an action potential, as in the left
panel of Fig 5e.
and the bottom window shows sodium and potassium currents in a
parallel fiber compartment during an action potential, as in the left
panel of Fig 5e.
 In the Genetic Setup Panel, clicking the button "GrC with FHF in All
Compartments" changes the model parameters by adding FHF modulation of
gNav to all axon and parallel fiber compartments and by adjusting gNav
and gKv to give a near-identical conducting spike waveform. By now
re-running the simulation, the parallel fiber spike-associated
currents now correspond to the right panel of Fig 5e
In the Genetic Setup Panel, clicking the button "GrC with FHF in All
Compartments" changes the model parameters by adding FHF modulation of
gNav to all axon and parallel fiber compartments and by adjusting gNav
and gKv to give a near-identical conducting spike waveform. By now
re-running the simulation, the parallel fiber spike-associated
currents now correspond to the right panel of Fig 5e
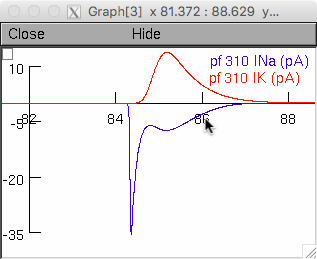 and show the consequently greater temporal overlap between INa and IK.
Usage instructions:
Auto-launch from ModelDB or download and extract the archive, then
under:
----
MSWIN
run mknrndll, cd to the archive and make the nrnmech.dll. Then double
click on the mosinit.hoc file.
----
MAC OS X
Drag and drop the GrC folder onto the mknrndll icon. Drag and drop
the mosinit.hoc file onto the nrngui icon.
----
Linux/Unix
Change directory to the GrC folder. run nrnivmodl.
Then type in terminal command line:
nrngui mosinit.hoc
----
Running the simulation will generate the simulations and plots shown
in Fig. 5 of the paper. The simulation starts automatically.
Questions on how to use this model should be directed to:
smgsolinas at gmail.com
or
goldfarb at genectr.hunter.cuny.edu
Changelog
---------
2022-12: Fix 9.0.0 upcoming error: used as both variable and function in files
Required for upcoming NEURON 9.0.0
and show the consequently greater temporal overlap between INa and IK.
Usage instructions:
Auto-launch from ModelDB or download and extract the archive, then
under:
----
MSWIN
run mknrndll, cd to the archive and make the nrnmech.dll. Then double
click on the mosinit.hoc file.
----
MAC OS X
Drag and drop the GrC folder onto the mknrndll icon. Drag and drop
the mosinit.hoc file onto the nrngui icon.
----
Linux/Unix
Change directory to the GrC folder. run nrnivmodl.
Then type in terminal command line:
nrngui mosinit.hoc
----
Running the simulation will generate the simulations and plots shown
in Fig. 5 of the paper. The simulation starts automatically.
Questions on how to use this model should be directed to:
smgsolinas at gmail.com
or
goldfarb at genectr.hunter.cuny.edu
Changelog
---------
2022-12: Fix 9.0.0 upcoming error: used as both variable and function in files
Required for upcoming NEURON 9.0.0
 the middle tier windows correspond to traces showing the first and the
ninth induced spike conducting along the axon and parallel fiber as in
Fig 5d,
the middle tier windows correspond to traces showing the first and the
ninth induced spike conducting along the axon and parallel fiber as in
Fig 5d,
 and the bottom window shows sodium and potassium currents in a
parallel fiber compartment during an action potential, as in the left
panel of Fig 5e.
and the bottom window shows sodium and potassium currents in a
parallel fiber compartment during an action potential, as in the left
panel of Fig 5e.
 In the Genetic Setup Panel, clicking the button "GrC with FHF in All
Compartments" changes the model parameters by adding FHF modulation of
gNav to all axon and parallel fiber compartments and by adjusting gNav
and gKv to give a near-identical conducting spike waveform. By now
re-running the simulation, the parallel fiber spike-associated
currents now correspond to the right panel of Fig 5e
In the Genetic Setup Panel, clicking the button "GrC with FHF in All
Compartments" changes the model parameters by adding FHF modulation of
gNav to all axon and parallel fiber compartments and by adjusting gNav
and gKv to give a near-identical conducting spike waveform. By now
re-running the simulation, the parallel fiber spike-associated
currents now correspond to the right panel of Fig 5e
 and show the consequently greater temporal overlap between INa and IK.
Usage instructions:
Auto-launch from ModelDB or download and extract the archive, then
under:
----
MSWIN
run mknrndll, cd to the archive and make the nrnmech.dll. Then double
click on the mosinit.hoc file.
----
MAC OS X
Drag and drop the GrC folder onto the mknrndll icon. Drag and drop
the mosinit.hoc file onto the nrngui icon.
----
Linux/Unix
Change directory to the GrC folder. run nrnivmodl.
Then type in terminal command line:
nrngui mosinit.hoc
----
Running the simulation will generate the simulations and plots shown
in Fig. 5 of the paper. The simulation starts automatically.
Questions on how to use this model should be directed to:
smgsolinas at gmail.com
or
goldfarb at genectr.hunter.cuny.edu
Changelog
---------
2022-12: Fix 9.0.0 upcoming error: used as both variable and function in files
Required for upcoming NEURON 9.0.0
and show the consequently greater temporal overlap between INa and IK.
Usage instructions:
Auto-launch from ModelDB or download and extract the archive, then
under:
----
MSWIN
run mknrndll, cd to the archive and make the nrnmech.dll. Then double
click on the mosinit.hoc file.
----
MAC OS X
Drag and drop the GrC folder onto the mknrndll icon. Drag and drop
the mosinit.hoc file onto the nrngui icon.
----
Linux/Unix
Change directory to the GrC folder. run nrnivmodl.
Then type in terminal command line:
nrngui mosinit.hoc
----
Running the simulation will generate the simulations and plots shown
in Fig. 5 of the paper. The simulation starts automatically.
Questions on how to use this model should be directed to:
smgsolinas at gmail.com
or
goldfarb at genectr.hunter.cuny.edu
Changelog
---------
2022-12: Fix 9.0.0 upcoming error: used as both variable and function in files
Required for upcoming NEURON 9.0.0