A Hodgkin Huxley model for conduction velocity in the medial giant
fiber of the earthworm, Lumbricus terrestris Heller and Crisp 2016
Model summary: The earthworm medial giant fiber (MGF) is composed of
many neurons electrically coupled by high fidelity gap junctions. In
addition, the MGF exhibits a distinct taper in diameter from anterior
to posterior. The role of these gap junctions and their interaction
with axonal taper in predicting conduction velocity has not been
studied closely in the annelid. A model of an electrical synapse in
the MGF was created to investigate the influence of, and interaction
between, these two parameters.
Model operation: In order to operate this model, open the .py file
containing the model code. Both the parameters for axon diameter (d)
and strength of gap junction coupling (gc) can be altered randomly
about some mean with some given standard deviation. These parameters
(mean and standard deviation) must be input by the user. Next, choose
how many trials you would like to test by changing the variable
"numTrials" at the beginning of the script. You can now run the
program.
In its default setting the model takes ten minutes or so to generate
this figure described below:
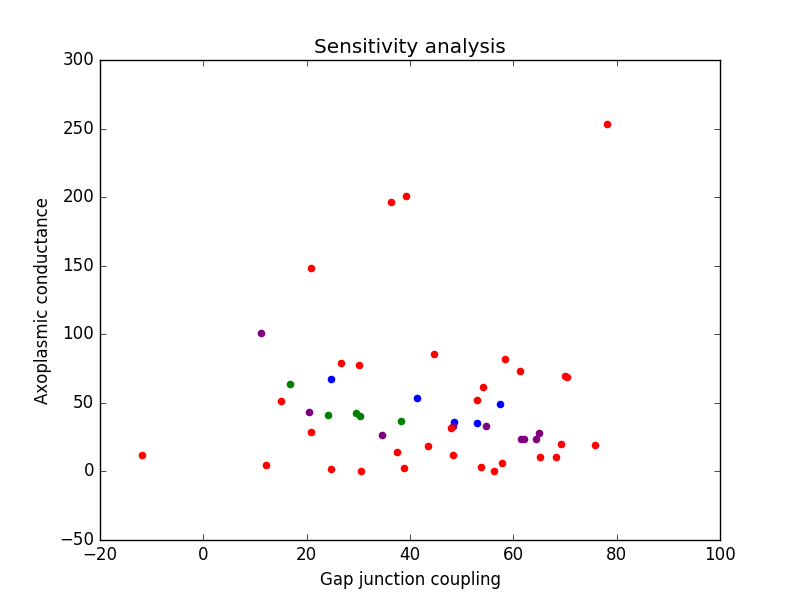 Model output: The model will output a scatter plot of succesful
combinations of gc (gap junction coupling strength) and ga (axonal
conductance). There will be four different categories of points
plotted. Red points correspond to failures: the combination of gc and
ga for that trial did not meet the physiological requirements. Blue
points correspond to fast conduction velocities: combinations of gc
and ga that give outputs similar to physiological results in the
anterior of the earthworm. Green points correspond to average
conduction velocities: combinations of ga and gc that give outputs
similar to physiological results in the middle of the earthworm
MGF. And finally, purple points correspond to slow conduction
velocities: combinations of ga and gc that give outputs similar to the
physiological results in the posterior of the earthworm MGF. In
addition, these sets of successful data points (the blue, green, and
purple points) will be output into three separate .csv files for
further analysis, if desired.
Model output: The model will output a scatter plot of succesful
combinations of gc (gap junction coupling strength) and ga (axonal
conductance). There will be four different categories of points
plotted. Red points correspond to failures: the combination of gc and
ga for that trial did not meet the physiological requirements. Blue
points correspond to fast conduction velocities: combinations of gc
and ga that give outputs similar to physiological results in the
anterior of the earthworm. Green points correspond to average
conduction velocities: combinations of ga and gc that give outputs
similar to physiological results in the middle of the earthworm
MGF. And finally, purple points correspond to slow conduction
velocities: combinations of ga and gc that give outputs similar to the
physiological results in the posterior of the earthworm MGF. In
addition, these sets of successful data points (the blue, green, and
purple points) will be output into three separate .csv files for
further analysis, if desired.
 Model output: The model will output a scatter plot of succesful
combinations of gc (gap junction coupling strength) and ga (axonal
conductance). There will be four different categories of points
plotted. Red points correspond to failures: the combination of gc and
ga for that trial did not meet the physiological requirements. Blue
points correspond to fast conduction velocities: combinations of gc
and ga that give outputs similar to physiological results in the
anterior of the earthworm. Green points correspond to average
conduction velocities: combinations of ga and gc that give outputs
similar to physiological results in the middle of the earthworm
MGF. And finally, purple points correspond to slow conduction
velocities: combinations of ga and gc that give outputs similar to the
physiological results in the posterior of the earthworm MGF. In
addition, these sets of successful data points (the blue, green, and
purple points) will be output into three separate .csv files for
further analysis, if desired.
Model output: The model will output a scatter plot of succesful
combinations of gc (gap junction coupling strength) and ga (axonal
conductance). There will be four different categories of points
plotted. Red points correspond to failures: the combination of gc and
ga for that trial did not meet the physiological requirements. Blue
points correspond to fast conduction velocities: combinations of gc
and ga that give outputs similar to physiological results in the
anterior of the earthworm. Green points correspond to average
conduction velocities: combinations of ga and gc that give outputs
similar to physiological results in the middle of the earthworm
MGF. And finally, purple points correspond to slow conduction
velocities: combinations of ga and gc that give outputs similar to the
physiological results in the posterior of the earthworm MGF. In
addition, these sets of successful data points (the blue, green, and
purple points) will be output into three separate .csv files for
further analysis, if desired.