This is the README for Purkinje cell model code for the paper:
Masoli, S., Solinas, S., & D’Angelo, E. (2015).
Action potential processing in a detailed Purkinje cell model reveals
a critical role for axonal compartmentalization. Frontiers in Cellular
Neuroscience, 9(February), 1–22.
http://doi.org/10.3389/fncel.2015.00047
Implementation done by Stefano Masoli in Python/Neuron.
The Purkinje cell (PC) is among the most complex neurons in the brain
and plays a critical role for cerebellar functioning. PCs operate as
fast pacemakers modulated by synaptic inputs but can switch from
simple spikes to complex bursts and, in some conditions, show
bistability. In contrast to original works emphasizing dendritic
Ca-dependent mechanisms, recent experiments have supported a primary
role for axonal Na-dependent processing, which could effectively
regulate spike generation and transmission to deep cerebellar nuclei
(DCN). In order to account for the numerous ionic mechanisms involved
(at present including Nav1.6, Cav2.1, Cav3.1, Cav3.2, Cav3.3, Kv1.1,
Kv1.5, Kv3.3, Kv3.4, Kv4.3, KCa1.1, KCa2.2, KCa3.1, Kir2.x, HCN1), we
have elaborated a multicompartmental model incorporating available
knowledge on localization and gating of PC ionic channels. The axon,
including initial segment (AIS) and Ranvier nodes (RNs), proved
critical to obtain appropriate pacemaking and firing frequency
modulation. Simple spikes initiated in the AIS and protracted
discharges were stabilized in the soma through Na-dependent
mechanisms, while somato-dendritic Ca channels contributed to sustain
pacemaking and to generate complex bursting at high discharge
regimes. Bistability occurred only following Na and Ca channel
down-regulation. In addition, specific properties in RNs K currents
were required to limit spike transmission frequency along the
axon. The model showed how organized electroresponsive functions could
emerge from the molecular complexity of PCs and showed that the axon
is fundamental to complement ionic channel compartmentalization
enabling action potential processing and transmission of specific
spike patterns to DCN.
Requirement:
The model was built under Mint (ubuntu based) with NEURON 7.3 and Python 2.7.
CHANGELOG - April 2021
The code runs with NEURON 7.8, NEURON 8 and with Python3.6 to 3.8
It requires a powerfull CPU with as many cores as possibile.
The model uses NEURON multisplit to distribute automatically the
calculation on all the available cores.
A typical 5s simulation takes about 15m on an AMD 8350 8 core CPU and
less than 4m on an AMD 1800x 8cores/16thread CPU.
Usage instructions:
Download and extract the archive.
Under Linux/Unix:
Change directory to "purkinjecell" folder.
Run nrnivmodl ./mod_files to compile the mod files.
The model is provided with 6 protocols:
Run "nrngui -python ./protocols/01_no_channels_ais_py3.py" for the first protocol.
The other protocols are numbered from 02 to 06.
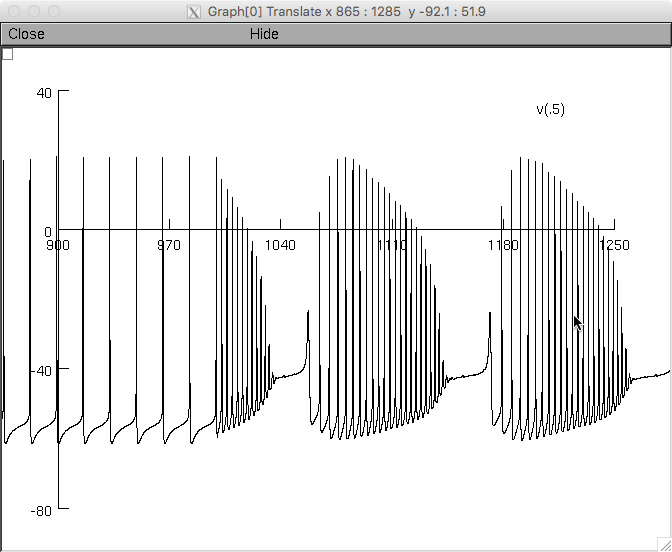
Here is a screenshot from running the command:
nrngui -python protocols/05_calcium_sodium_bursts_py3.py
 Attention:
The model does not work with the variable time step!
If you would like more help please refer to:
https://senselab.med.yale.edu/ModelDB/NEURON_DwnldGuide.cshtml
NOTES:
Not tested under Win installations.
Attention:
The model does not work with the variable time step!
If you would like more help please refer to:
https://senselab.med.yale.edu/ModelDB/NEURON_DwnldGuide.cshtml
NOTES:
Not tested under Win installations.
 Attention:
The model does not work with the variable time step!
If you would like more help please refer to:
https://senselab.med.yale.edu/ModelDB/NEURON_DwnldGuide.cshtml
NOTES:
Not tested under Win installations.
Attention:
The model does not work with the variable time step!
If you would like more help please refer to:
https://senselab.med.yale.edu/ModelDB/NEURON_DwnldGuide.cshtml
NOTES:
Not tested under Win installations.
Attention: The model does not work with the variable time step! If you would like more help please refer to: https://senselab.med.yale.edu/ModelDB/NEURON_DwnldGuide.cshtml NOTES: Not tested under Win installations.