//========================================================================================================================
C++ CODE FOR IMPLEMENTING AN UNIFIED THALAMIC MODEL FOR THE GENERATION OF MULTIPLE DISTINCT OSCILLATIONS
Guoshi Li
Department of Psychiatry
Univeristy of North Carolina at Chapel Hill
Chapel Hill, NC 27599
A full description of the model can be found in:
Guoshi Li, Craig Henriquez and Flavio Frohlich (2017) Unified Thalamic Model Generates Multiple Distinct Oscillations with State-dependent Entrainment by Stimulation. PLOS Computational Biology.
http://dx.doi.org/10.1371/journal.pcbi.1005797
For questions, please email: guoshi_li@med.unc.edu
//========================================================================================================================
Then thalamic model is implemented with C++ and simulation is run under Ubuntu Linux originally using the Eclipse program. A makefile was added so that you can type make to make the simulator in unix/linux.
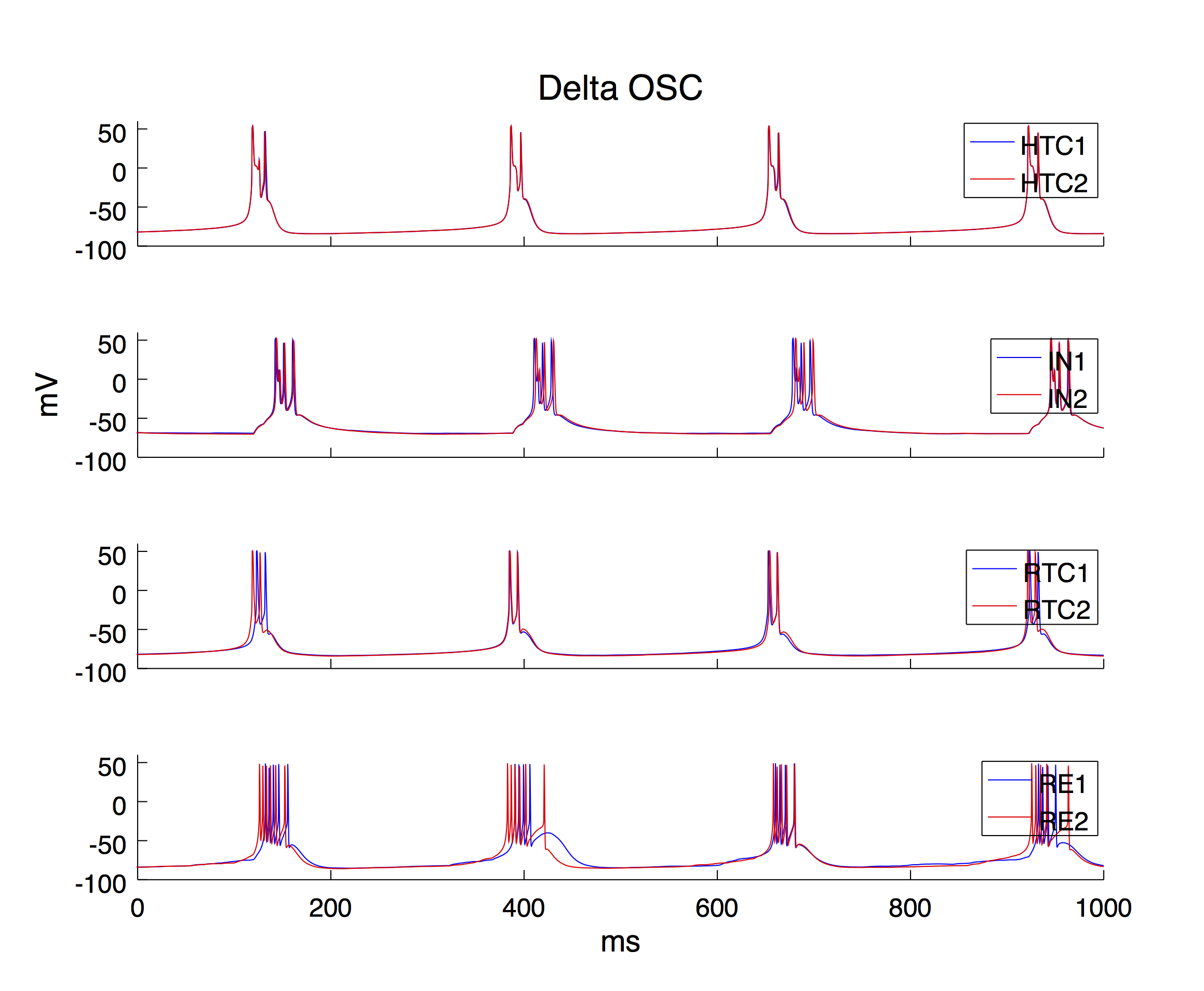
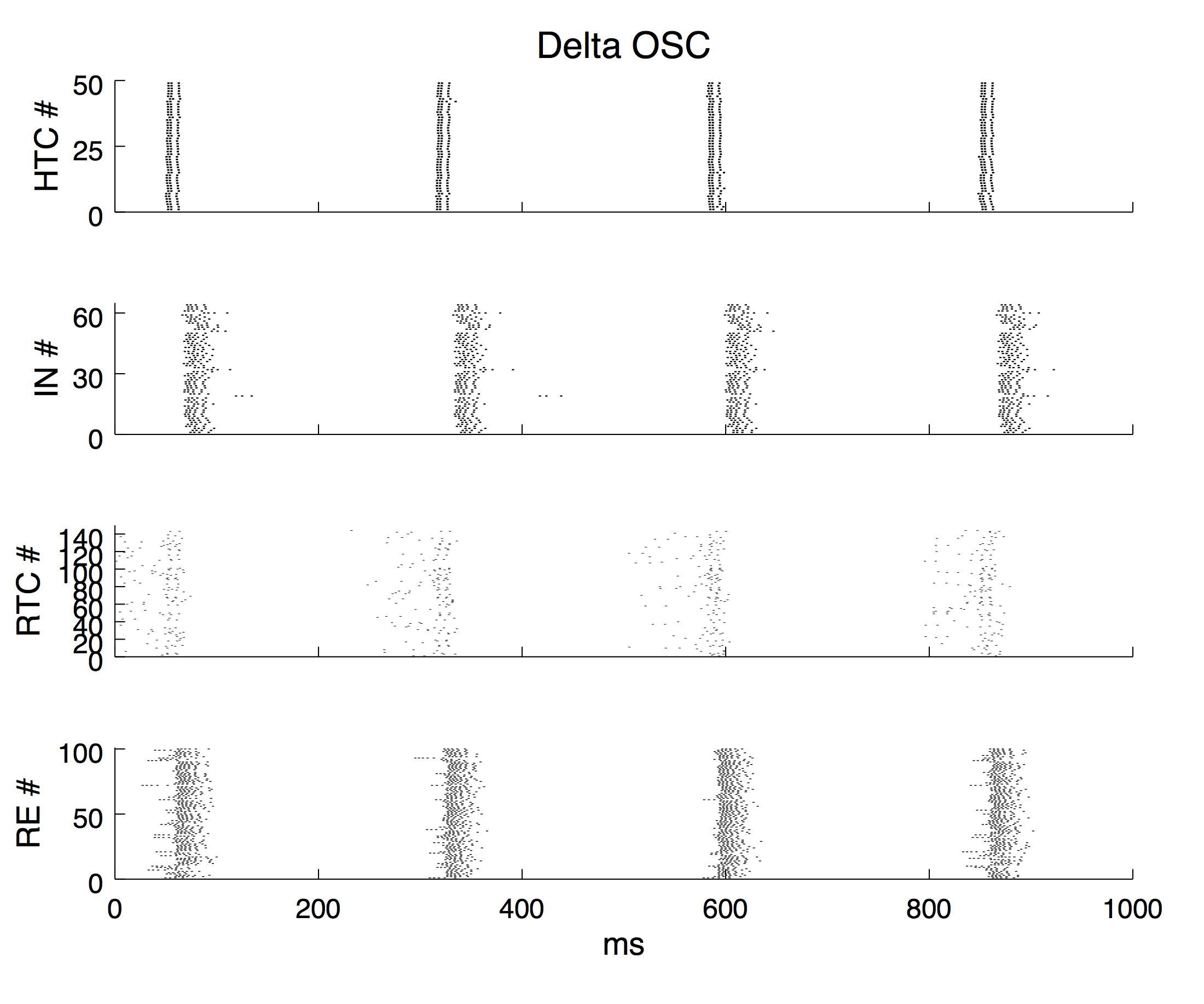
The thalamic network model contains 49 high-threshold bursting thalamocoritcal cells (HTCs), 144 regular relay-mode thalamocortical cells (RTCs), 64 local interneurons (INs) and 100 reticular cells (REs) each placed in a 2D grid.
The package contains two folders:
"src" folder contains the source code for ionic and synapitc channels
"data" folder to store data from network simulations
The main function is contained in Net.cpp.
Other major files are described as follows:
Constants.h: Specify the parameters of the thalamic model
TC.cpp: Define TC cell class
IN.cpp:Define IN cell class
RE.cpp:Define RE cell class
TC.h: Header file for TC cell object
IN.h: Header file for IN cell object
RE.h: Header file for RE cell object
The variable "OSC" defined in Constants.h determines which oscillation state is simulated:
2: Spindle oscillation
3: Alpha oscillaiton
4: Gamma oscillation
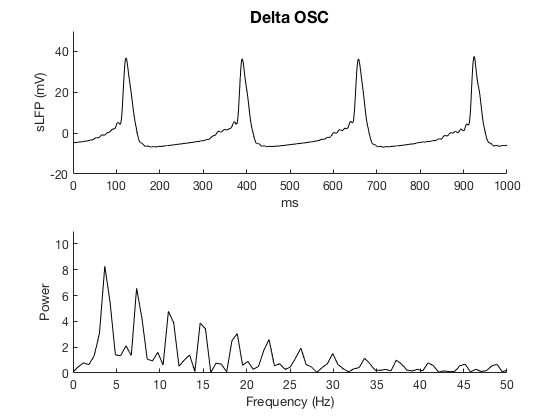
The simulation step is set to 0.02 ms and the default simulation time is 3000 ms (3 sec; set by the varialbe "t_end" in Constants.h). To generate the same Figure 2 as in the paper, the following simulation time is recommended:
Delta oscillaton: 3000 ms
Spindle oscillaton: 4000 ms
Alpha oscillaton: 2000 ms
Gamma oscillation: 2000 ms
//========================================================================================================================
The default simulation is without stimulation! To turn on stimulation to the LGN, set "FLAG_STIMULATION_LGN_PULSE" in Constants.h to 1 To turn on stimulation to the TRN, set "FLAG_STIMULATION_TRN_PULSE" in Constants.h to 1
When stimulation is on, pulsatile stimulus will be delivered to the LGN or TRN neurons and the stimulation frequency will increase from 1 Hz to 50 Hz (default). Each frequency step lasts for 1 second. Adjust the simulation time ("t_end") appropriately depending on the desired range of simulated frequency. To stimulate the network from 1 to 50 Hz, the simulation time needs to be 52,000 ms (52 seconds).
20171109 Model updated to include a makefile and screenshots.