This directory contains the Neuron source code for the cortical Layer 5 pyramidal cell model used in:
Anderson R, Farokhniaee A, Gunalan K., Howell B, McIntyre C., Action potential initiation, propagation, and cortical invasion in the hyperdirect pathway during subthalamic deep brain stimulation, Brain Stimulation (2018), https://doi.org/10.1016/j.brs.2018.05.008
Parts of this model are based on the following works:
Distinct Contributions of Na(V)1.6 and Na(V)1.2 in Action Potential Initiation and Backpropagation Wenqin Hu, Cuiping Tian, Tun Li, Mingpo Yang, Han Hou & Yousheng Shu (2009) Nat Neurosci 12(8): 996-1002.
Mainen, Z. F. and Sejnowski, T. J. Nature 382: 363-6 (1996) Yu, Y., Shu, Y., et al. J Neurosci 28: 7260-72 (2008) Shu, Y., Hasenstaub, A., et al. Nature 441: 761-5. (2006)
Shu Y, Duque A, Yu Y, Haider B, McCormick DA. 2007. Properties of action-potential initiation in neocortical pyramidal cells: evidence from whole cell axon recordings. J Neurophysiol. 97(1):746-60.
The 3D structure of the cortical axon arbor is from:
Binzegger T, Douglas RJ, Martin KA. 2005. Axons in the cat visual cortex are topologically self-similar. Cereb Cortex. 15(2):152-65.
The hyperdirect collateral structure is adapted from:
Kita T, Kita H. 2012. The subthalamic nucleus is one of multiple innervation site for long-range corticofugal axons: a single-axon tracing study in the rat. J Neurosci. 32(17):5990-9.
BRIEF OVERVIEW OF THE CONTENTS
The model included is used to reproduce the results seen in Figures 2-7 of the paper. Due to the complexity of implementation of this model, it has been configured to reproduce the contents of Figure 3A and 3B. Therefore, the model will record the membrane potentials along the complete neuron during 1 extracellularly evoked antidromic stimulation event and write these values to the file created in the directory membraneVoltages. The 3D potential can then be graphed utilizing the Matlab code found in the directory Matlab using the file shapePlotV2.m. Sample data is included to aid in the process. Code and comments are given to recreate the results in the remaining figures but the code is not explicitly set up to recreate all figures..
Directories are organized as follows:
binzeggerAxon: The location of the structure for the cortical axon arbor, the hyperdirect collateral and the Shu membrane biophysics for the myelinated structure.
experiment: The main control file (Pyramidal_Main.hoc, adapted from Hu et al.), the excitatory/inhibitory input control variables and control files to do membrane voltage measurement and AP counting.
extracellular: The extracellular stimulation computed potentials.
lib: Density mechanism definition
Matlab: Matlab code for plotting the membrane potentials in the shape plot
mechanism: The channel mechanism module files.
membraneVoltages: Saved membrane voltages for the 3D shape plot.
morphology: original morphology definition from Hu
mosinit.hoc: The mosinit file.
Readme.html: This readme.
session: Session file.
USAGE
- If needed, first compile the module files located in the the directory MECHANISM by running the command:
nrnivmodl mechanism/*.mod
- To run the model, execute the file mosinit.hoc
- Most configuration occurs in the file experiment/Pyramidal_Main.hoc
After the simulation completes runnning the matlab program by typing the name shapePlotV2 on the command line and then entering times that are 1 ms ahead of the times listed in Fig3A will generate figures from the paper:
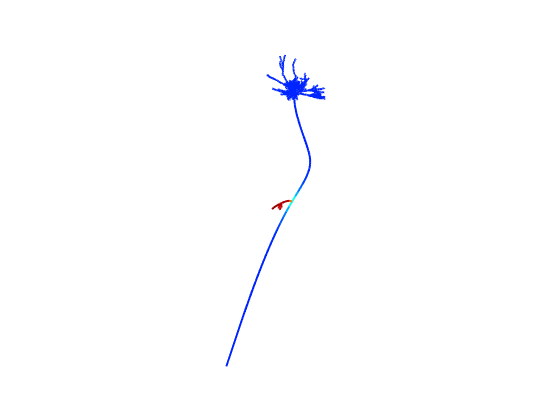 0 ms
0 ms
 1 ms
1 ms
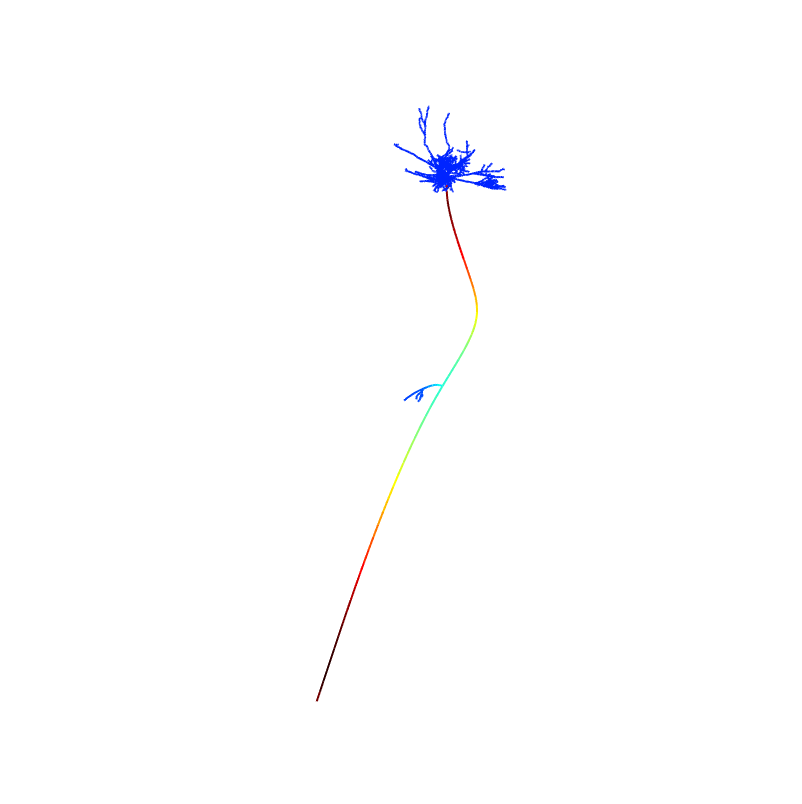 2 ms
2 ms
For example entering 2.5 will show a time between the papers fig 3A 1 and 2ms images:
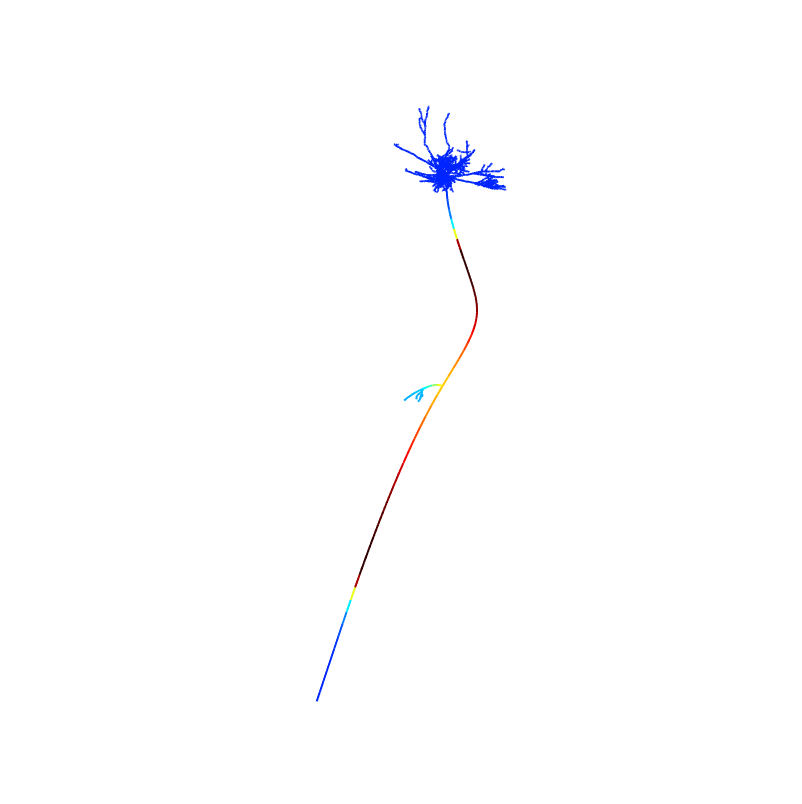
 0 ms
0 ms 1 ms
1 ms 2 ms
2 ms