Vasalou C, Henson MA (2010) A multiscale model to investigate circadian rhythmicity of pacemaker neurons in the suprachiasmatic nucleus. PLoS Comput Biol 6:e1000706 [PubMed]
are available at the cellml.org website (translated from SBML from biomodels): https://models.cellml.org/e/25/vasalou_2010.cellml/view Example: To recreate a few traces in opencor from figure 2 in the paper, change the ending point to 100 hours and the interval point to 0.1 hours. The 100 hours lets the model stabilize. Create a few graphs (use "+" sign) and then populate the graphs by selecting a graph by clicking on it, and adding to one of each by right-clicking IK, ICa, and fr (firing rate) under the membrane list, and select Plot against integration variable. Running (click upper left play triangle) creates the following graph contain a few traces from figure 2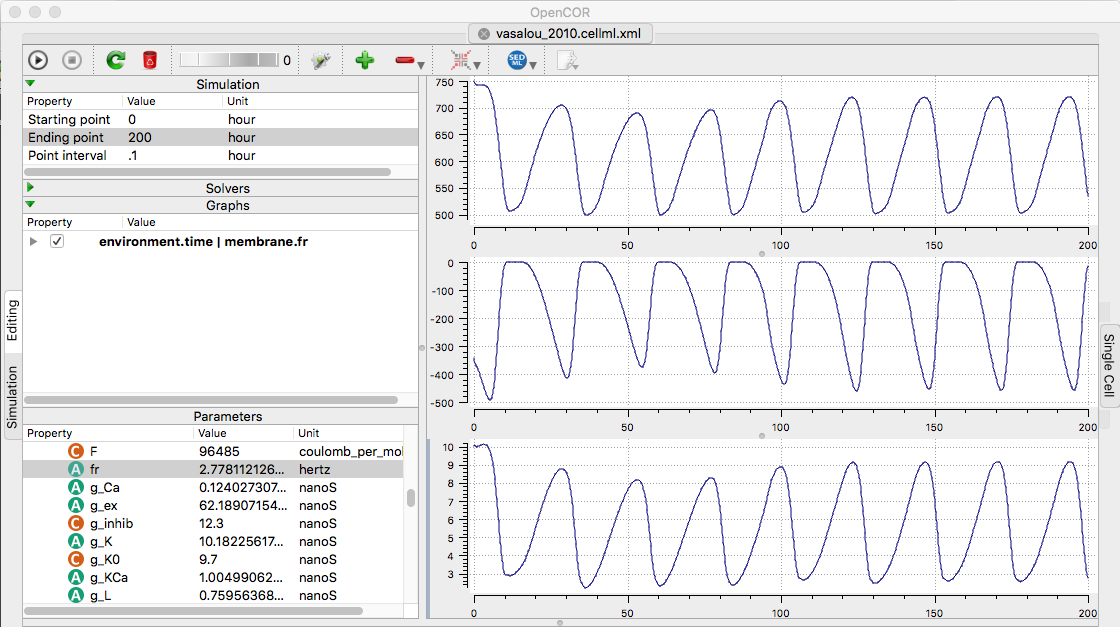 (Above traces are IK, ICa, and firing rate respectively plotted vs hours. Each cycle is 24 hours.)
(Above traces are IK, ICa, and firing rate respectively plotted vs hours. Each cycle is 24 hours.)