Representative Simulations for
Wang, X.-J. and Buzsaki G. (1996) Gamma oscillations by synaptic
inhibition in a hippocampal interneuronal network. J. Neurosci. 16,
6402-6413.
Download archive and compile the mod files (mknrndll in mswin and mac,
nrnivmodl in linux/unix) and begin by starting mosinit.hoc (double
click in mswin, drag and drop onto nrngui on mac, or "nrngui
mosinit.hoc" in linux/unix). Press either figure button to generate:
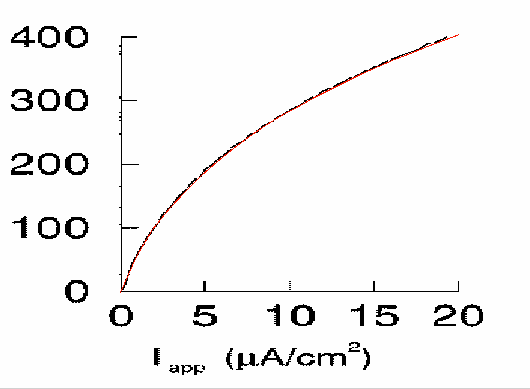 Fig. 1a
Small discrepancies between gif background in Grapher and the
simulation is probably due to slightly incorrect scaling and origin of
the gif.
Fig. 1a
Small discrepancies between gif background in Grapher and the
simulation is probably due to slightly incorrect scaling and origin of
the gif.
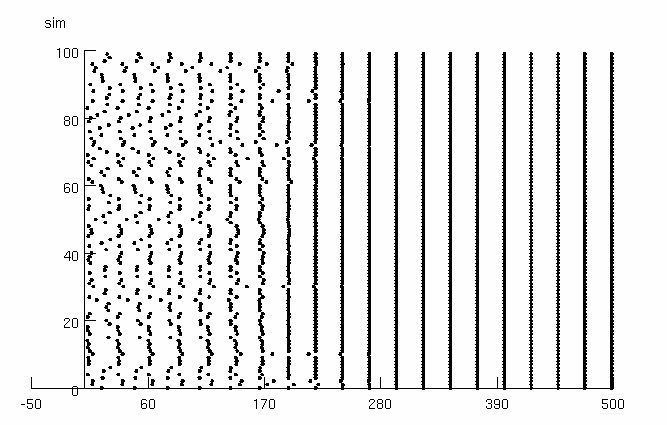 Default parameters replicate Fig. 3A (phi0=5; current injection amp0=1
muA/cm2).
Default parameters replicate Fig. 3A (phi0=5; current injection amp0=1
muA/cm2).
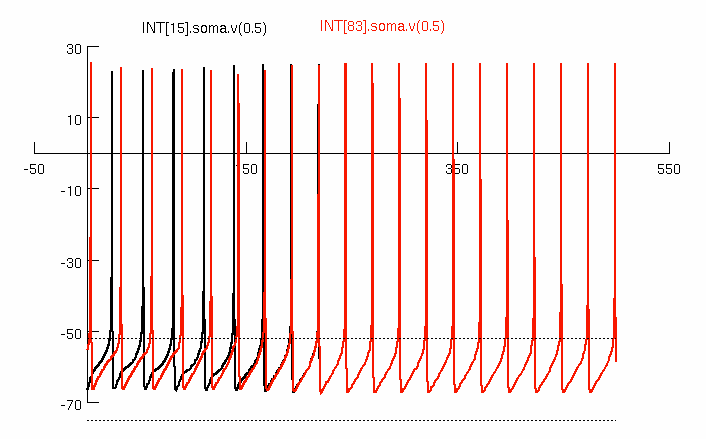 Fig. 3B parameters are (phi0=3.3; amp0=1.2); 3C (phi0=2.0; amp0=1.4).
Note: this simulation takes a little more than 3 minutes on a 2GHz
linux machine, under a minute on a 2012 MacBook Pro (laptop).
Start the simulation by pressing the Run button or the Init&Run
button. When the simulation stops, press the "Spike plot" button and
the "Plot cells A&B" button.
Parameter control panel permits change in parameters as follows:
phi phi0
Current inj. amp0
Run redundant with run button on run control
Randomize rerandomizes initial conditions (Chosen as a uniform
distribution between -68 and -55 mV; this range was
chosen to give sync at approximately the cycle seen in
the paper.)
Spike Plot Put up a graph of spike times (Note that you can stop
simulation, put up a graph, and restart. This graph
is only a snapshot and will not be updated.)
Plot cells A&B Overlay 2 cells' voltage plots. Cell # chosen below
must be between 0 and 99. Horizontal lines are drawn
at -52 and -75 mV to compare with right-side graphs of
Fig. 3 in paper.
Cell A Choose cell to plot.
Cell B Choose other cell to plot.
Technical notes:
In the original paper, current injection is given in units of mA/cm2.
Here we have chosen a cell size of 100um/cm2 so that nA is equivalent
to mA/cm2.
In the original paper, phi directly alters the time constant as a
factor in the differential equation. In this implementation,
phi(phi0) alters celsius appropriately.
Bill Lytton developed the fig 3a simulation and model
descriptions. Michael Hines created the fig1a gui.
Changelog
---------
20140821 ModelDB admininstrator: updates to run on mswin
20220520 Updated MOD files to contain valid C++ and be compatible
with the upcoming versions 8.2 and 9.0 of NEURON.
Fig. 3B parameters are (phi0=3.3; amp0=1.2); 3C (phi0=2.0; amp0=1.4).
Note: this simulation takes a little more than 3 minutes on a 2GHz
linux machine, under a minute on a 2012 MacBook Pro (laptop).
Start the simulation by pressing the Run button or the Init&Run
button. When the simulation stops, press the "Spike plot" button and
the "Plot cells A&B" button.
Parameter control panel permits change in parameters as follows:
phi phi0
Current inj. amp0
Run redundant with run button on run control
Randomize rerandomizes initial conditions (Chosen as a uniform
distribution between -68 and -55 mV; this range was
chosen to give sync at approximately the cycle seen in
the paper.)
Spike Plot Put up a graph of spike times (Note that you can stop
simulation, put up a graph, and restart. This graph
is only a snapshot and will not be updated.)
Plot cells A&B Overlay 2 cells' voltage plots. Cell # chosen below
must be between 0 and 99. Horizontal lines are drawn
at -52 and -75 mV to compare with right-side graphs of
Fig. 3 in paper.
Cell A Choose cell to plot.
Cell B Choose other cell to plot.
Technical notes:
In the original paper, current injection is given in units of mA/cm2.
Here we have chosen a cell size of 100um/cm2 so that nA is equivalent
to mA/cm2.
In the original paper, phi directly alters the time constant as a
factor in the differential equation. In this implementation,
phi(phi0) alters celsius appropriately.
Bill Lytton developed the fig 3a simulation and model
descriptions. Michael Hines created the fig1a gui.
Changelog
---------
20140821 ModelDB admininstrator: updates to run on mswin
20220520 Updated MOD files to contain valid C++ and be compatible
with the upcoming versions 8.2 and 9.0 of NEURON.
 Fig. 1a
Small discrepancies between gif background in Grapher and the
simulation is probably due to slightly incorrect scaling and origin of
the gif.
Fig. 1a
Small discrepancies between gif background in Grapher and the
simulation is probably due to slightly incorrect scaling and origin of
the gif.
 Default parameters replicate Fig. 3A (phi0=5; current injection amp0=1
muA/cm2).
Default parameters replicate Fig. 3A (phi0=5; current injection amp0=1
muA/cm2).
 Fig. 3B parameters are (phi0=3.3; amp0=1.2); 3C (phi0=2.0; amp0=1.4).
Note: this simulation takes a little more than 3 minutes on a 2GHz
linux machine, under a minute on a 2012 MacBook Pro (laptop).
Start the simulation by pressing the Run button or the Init&Run
button. When the simulation stops, press the "Spike plot" button and
the "Plot cells A&B" button.
Parameter control panel permits change in parameters as follows:
phi phi0
Current inj. amp0
Run redundant with run button on run control
Randomize rerandomizes initial conditions (Chosen as a uniform
distribution between -68 and -55 mV; this range was
chosen to give sync at approximately the cycle seen in
the paper.)
Spike Plot Put up a graph of spike times (Note that you can stop
simulation, put up a graph, and restart. This graph
is only a snapshot and will not be updated.)
Plot cells A&B Overlay 2 cells' voltage plots. Cell # chosen below
must be between 0 and 99. Horizontal lines are drawn
at -52 and -75 mV to compare with right-side graphs of
Fig. 3 in paper.
Cell A Choose cell to plot.
Cell B Choose other cell to plot.
Technical notes:
In the original paper, current injection is given in units of mA/cm2.
Here we have chosen a cell size of 100um/cm2 so that nA is equivalent
to mA/cm2.
In the original paper, phi directly alters the time constant as a
factor in the differential equation. In this implementation,
phi(phi0) alters celsius appropriately.
Bill Lytton developed the fig 3a simulation and model
descriptions. Michael Hines created the fig1a gui.
Changelog
---------
20140821 ModelDB admininstrator: updates to run on mswin
20220520 Updated MOD files to contain valid C++ and be compatible
with the upcoming versions 8.2 and 9.0 of NEURON.
Fig. 3B parameters are (phi0=3.3; amp0=1.2); 3C (phi0=2.0; amp0=1.4).
Note: this simulation takes a little more than 3 minutes on a 2GHz
linux machine, under a minute on a 2012 MacBook Pro (laptop).
Start the simulation by pressing the Run button or the Init&Run
button. When the simulation stops, press the "Spike plot" button and
the "Plot cells A&B" button.
Parameter control panel permits change in parameters as follows:
phi phi0
Current inj. amp0
Run redundant with run button on run control
Randomize rerandomizes initial conditions (Chosen as a uniform
distribution between -68 and -55 mV; this range was
chosen to give sync at approximately the cycle seen in
the paper.)
Spike Plot Put up a graph of spike times (Note that you can stop
simulation, put up a graph, and restart. This graph
is only a snapshot and will not be updated.)
Plot cells A&B Overlay 2 cells' voltage plots. Cell # chosen below
must be between 0 and 99. Horizontal lines are drawn
at -52 and -75 mV to compare with right-side graphs of
Fig. 3 in paper.
Cell A Choose cell to plot.
Cell B Choose other cell to plot.
Technical notes:
In the original paper, current injection is given in units of mA/cm2.
Here we have chosen a cell size of 100um/cm2 so that nA is equivalent
to mA/cm2.
In the original paper, phi directly alters the time constant as a
factor in the differential equation. In this implementation,
phi(phi0) alters celsius appropriately.
Bill Lytton developed the fig 3a simulation and model
descriptions. Michael Hines created the fig1a gui.
Changelog
---------
20140821 ModelDB admininstrator: updates to run on mswin
20220520 Updated MOD files to contain valid C++ and be compatible
with the upcoming versions 8.2 and 9.0 of NEURON.