These simulations accompany the publication:
Schmidt-Hieber C, Jonas P, Bischofberger J (2007)
Subthreshold Dendritic Signal Processing and Coincidence Detection
in Dentate Gyrus Granule Cells. J Neurosci 27:8430-8441
For installation instructions, see INSTALL.txt
Simulations have been tested using NEURON 5.9 and 6.0 under
Windows and GNU/Linux. The gnuplot script has been tested with
gnuplot 4.2 (http://www.gnuplot.info).
Once the simulation starts you can click on buttons in the gui
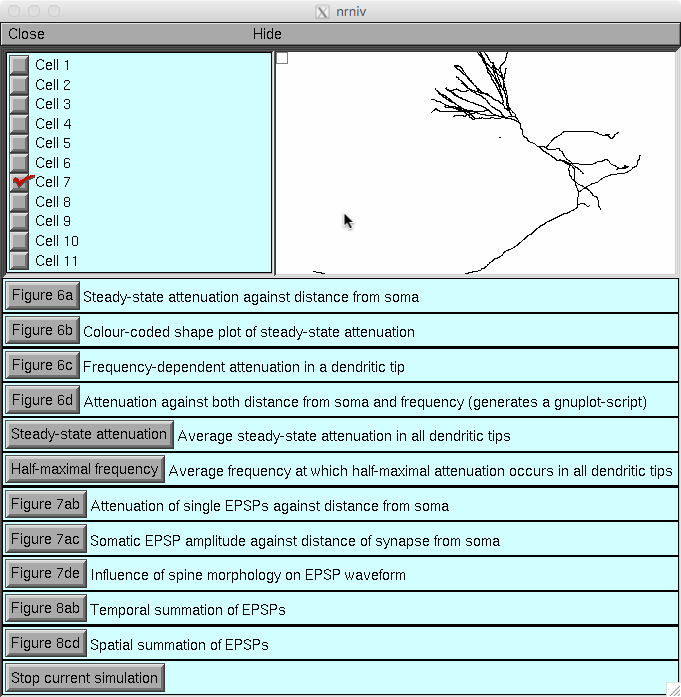 to generate selected figures in the paper:
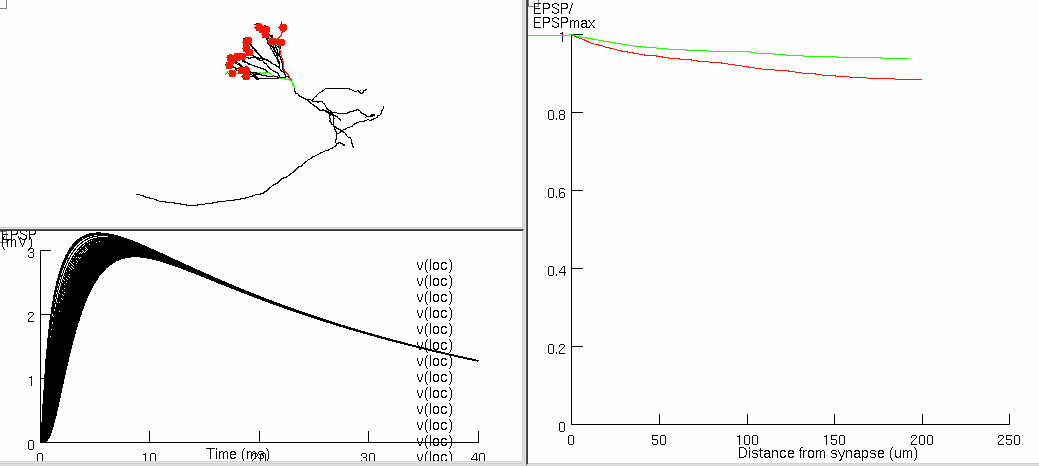
Figure 6a
to generate selected figures in the paper:
Figure 6a
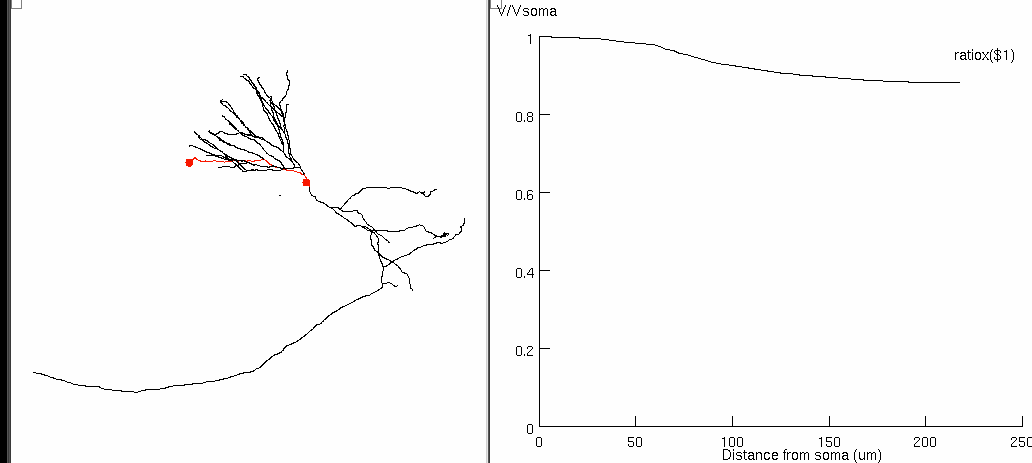 Figure 6b
Figure 6b
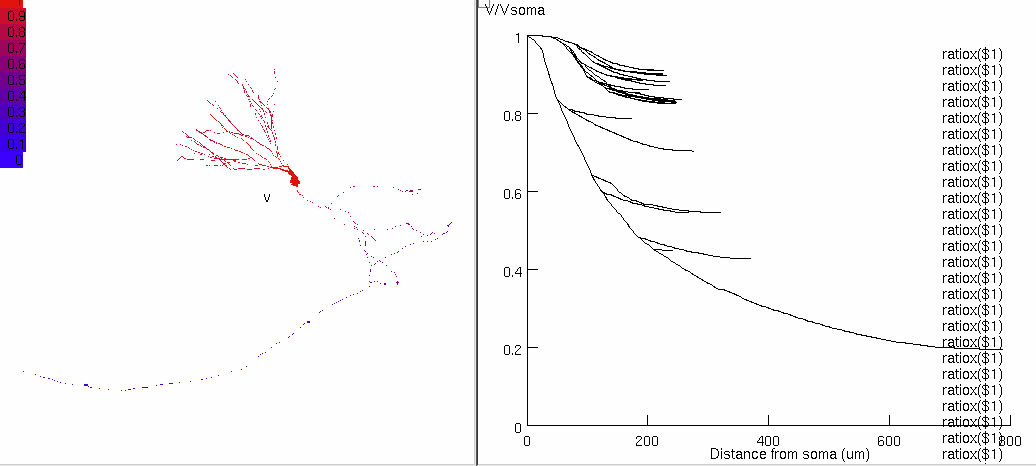 Figure 6c
Figure 6c
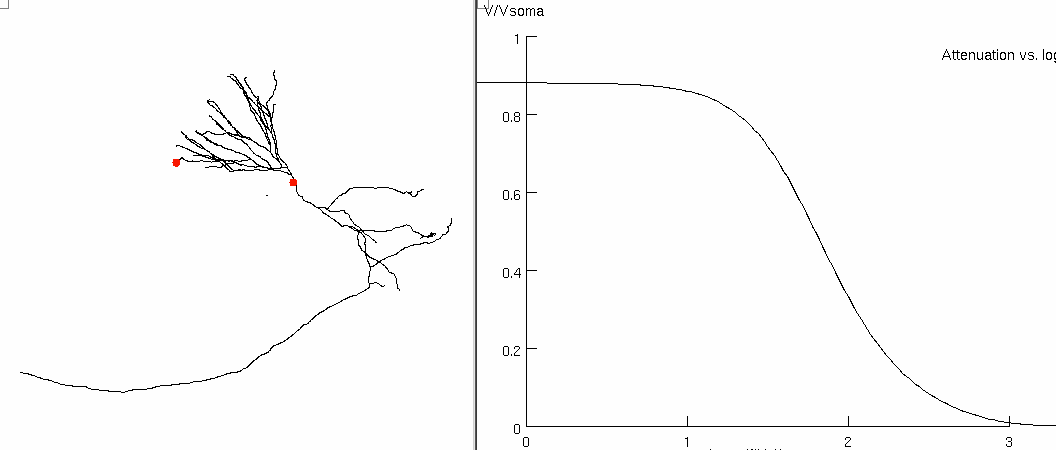 Figure 7ab
Figure 7ab
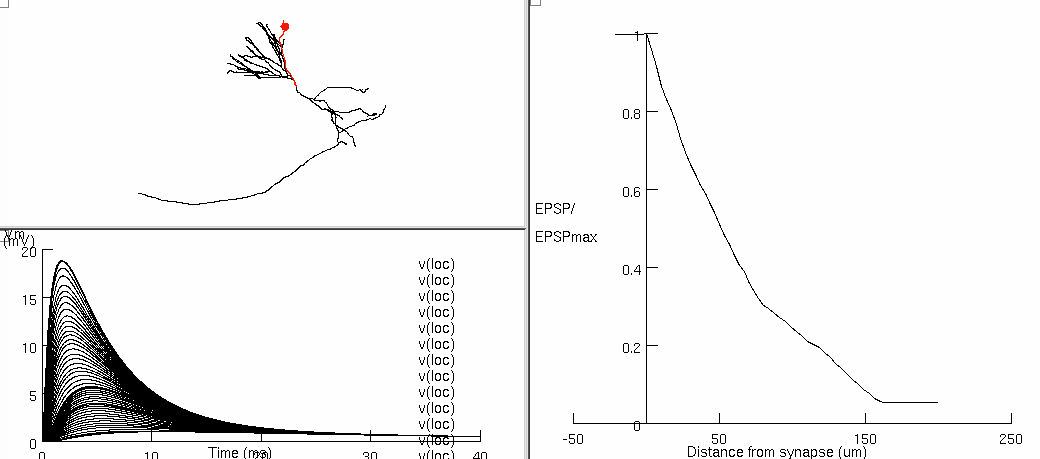 Figure 7ac
Figure 7ac
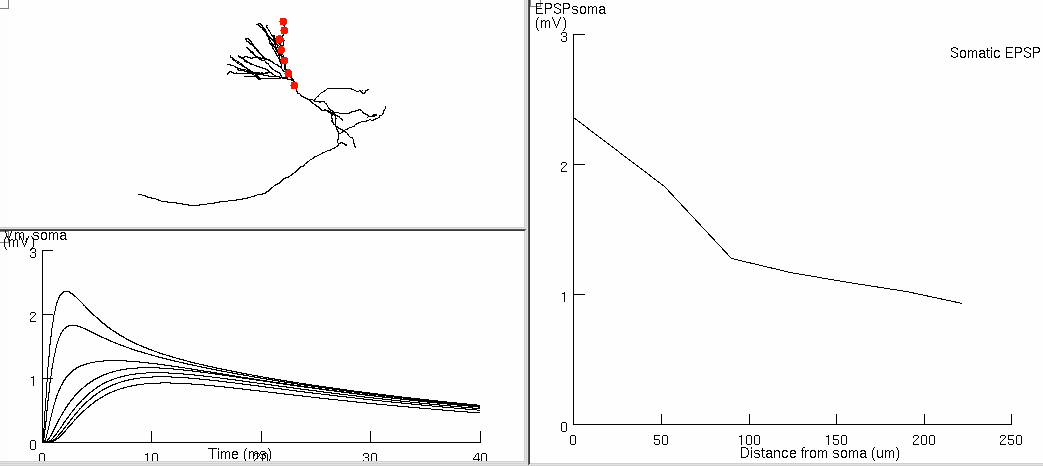 Figure 7d
Figure 7d
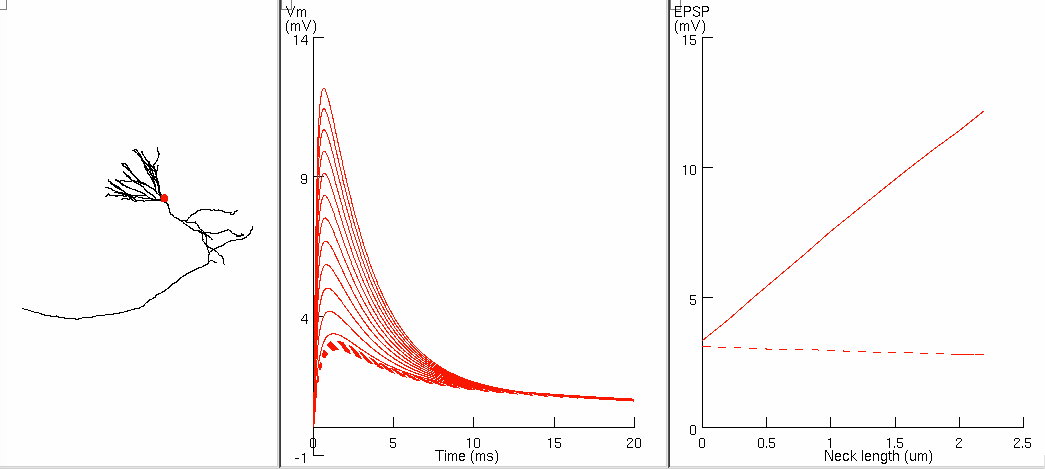 Figure 8ab
Figure 8ab
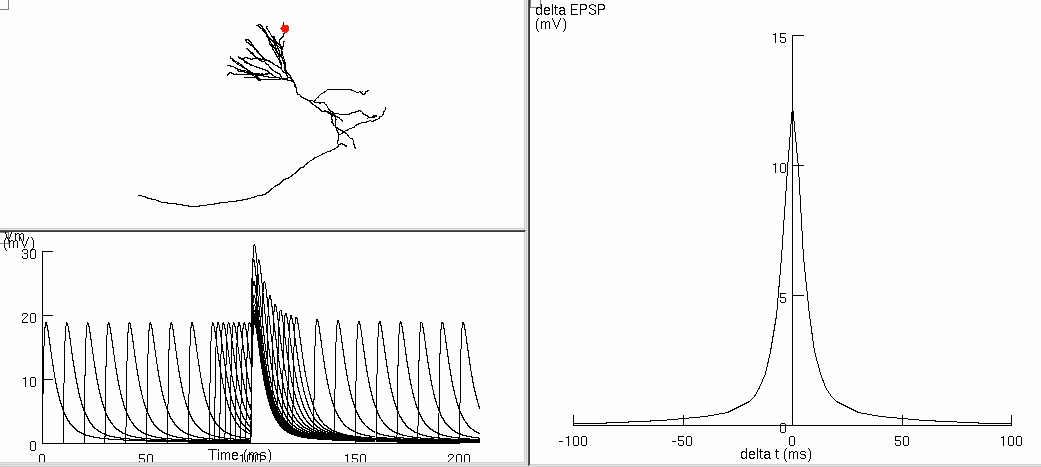 Figure 8cd
Figure 8cd
 Send bug reports and comments to
christoph.schmidt-hieber at uni-freiburg.de
Description of files:
./gc.hoc Starts the simulations. Requires mod files to be compiled.
./asin.mod arcsine function, forwarded to hoc from math.h
./spines.mod Introduces spine area scale factor as a range variable
(scale_spines).
Optionally, spine counts can be used as a range variable
as well (count_spines).
./synampa.mod Synaptic conductance change with a biexponential time course.
./share/calcSpines.hoc Calculate spine area scale factor from spine counts. Corrects
for spines hidden behind the shaft in 2D projections.
./share/config.hoc Simulation configuration settings.
./share/controlpanel.hoc Main control panel allowing cell and figure selection.
./share/figures.hoc Procedures reproducing figures.
./share/fixnseg.hoc d_lambda rule implementation.
./share/genutils.hoc Some general-purpose routines.
./share/spine.hoc Creates a spine, consisting of a neck (spine[0]) and a
head (spine[1])
./share/template.plt Template script file for creating a gnuplot script that
will show a 3D plot as in figure 6D.
./cell_x/membrane.hoc Loads cell morphology and connections, initializes spines,
inserts passive membrane properties. Uses a class "cell_x"
so that you can easily toggle between cells or use multiple
objects for network simulations.
./cell_x/morpho.hoc Loads morphology and spines without making use of a class.
Intended for standalone use of cells.
./cell_x/morpho.txt Coordinates and diameters of 3D points as plain text. Needed
by membrane.hoc to load 3D morphology from a procedure.
20071005
Cell templates now adhere to the "NetworkReadyCell" syntax - fixed some minor bugs. Schmidt-Hieber.
20170328
* Added a new cell (cell_9) and an artificial cell (cell_10) that were not
part of the original 2007 publication.
* Added CA1 PC model (cell_11) for consistency. See
Golding NL, Mickus TJ, Katz Y, Kath WL, Spruston N (2005)
Factors mediating powerful voltage attenuation along CA1 pyramidal neuron
dendrites. J Physiol 568:69-82.
To run the original simulations as shown in the paper, use the original model from
https://www.janelia.org/sites/default/files/Attenuation.zip
Send bug reports and comments to
christoph.schmidt-hieber at uni-freiburg.de
Description of files:
./gc.hoc Starts the simulations. Requires mod files to be compiled.
./asin.mod arcsine function, forwarded to hoc from math.h
./spines.mod Introduces spine area scale factor as a range variable
(scale_spines).
Optionally, spine counts can be used as a range variable
as well (count_spines).
./synampa.mod Synaptic conductance change with a biexponential time course.
./share/calcSpines.hoc Calculate spine area scale factor from spine counts. Corrects
for spines hidden behind the shaft in 2D projections.
./share/config.hoc Simulation configuration settings.
./share/controlpanel.hoc Main control panel allowing cell and figure selection.
./share/figures.hoc Procedures reproducing figures.
./share/fixnseg.hoc d_lambda rule implementation.
./share/genutils.hoc Some general-purpose routines.
./share/spine.hoc Creates a spine, consisting of a neck (spine[0]) and a
head (spine[1])
./share/template.plt Template script file for creating a gnuplot script that
will show a 3D plot as in figure 6D.
./cell_x/membrane.hoc Loads cell morphology and connections, initializes spines,
inserts passive membrane properties. Uses a class "cell_x"
so that you can easily toggle between cells or use multiple
objects for network simulations.
./cell_x/morpho.hoc Loads morphology and spines without making use of a class.
Intended for standalone use of cells.
./cell_x/morpho.txt Coordinates and diameters of 3D points as plain text. Needed
by membrane.hoc to load 3D morphology from a procedure.
20071005
Cell templates now adhere to the "NetworkReadyCell" syntax - fixed some minor bugs. Schmidt-Hieber.
20170328
* Added a new cell (cell_9) and an artificial cell (cell_10) that were not
part of the original 2007 publication.
* Added CA1 PC model (cell_11) for consistency. See
Golding NL, Mickus TJ, Katz Y, Kath WL, Spruston N (2005)
Factors mediating powerful voltage attenuation along CA1 pyramidal neuron
dendrites. J Physiol 568:69-82.
To run the original simulations as shown in the paper, use the original model from
https://www.janelia.org/sites/default/files/Attenuation.zip
 to generate selected figures in the paper:
Figure 6a
to generate selected figures in the paper:
Figure 6a
 Figure 6b
Figure 6b
 Figure 6c
Figure 6c
 Figure 7ab
Figure 7ab
 Figure 7ac
Figure 7ac
 Figure 7d
Figure 7d
 Figure 8ab
Figure 8ab
 Figure 8cd
Figure 8cd
 Send bug reports and comments to
christoph.schmidt-hieber at uni-freiburg.de
Description of files:
./gc.hoc Starts the simulations. Requires mod files to be compiled.
./asin.mod arcsine function, forwarded to hoc from math.h
./spines.mod Introduces spine area scale factor as a range variable
(scale_spines).
Optionally, spine counts can be used as a range variable
as well (count_spines).
./synampa.mod Synaptic conductance change with a biexponential time course.
./share/calcSpines.hoc Calculate spine area scale factor from spine counts. Corrects
for spines hidden behind the shaft in 2D projections.
./share/config.hoc Simulation configuration settings.
./share/controlpanel.hoc Main control panel allowing cell and figure selection.
./share/figures.hoc Procedures reproducing figures.
./share/fixnseg.hoc d_lambda rule implementation.
./share/genutils.hoc Some general-purpose routines.
./share/spine.hoc Creates a spine, consisting of a neck (spine[0]) and a
head (spine[1])
./share/template.plt Template script file for creating a gnuplot script that
will show a 3D plot as in figure 6D.
./cell_x/membrane.hoc Loads cell morphology and connections, initializes spines,
inserts passive membrane properties. Uses a class "cell_x"
so that you can easily toggle between cells or use multiple
objects for network simulations.
./cell_x/morpho.hoc Loads morphology and spines without making use of a class.
Intended for standalone use of cells.
./cell_x/morpho.txt Coordinates and diameters of 3D points as plain text. Needed
by membrane.hoc to load 3D morphology from a procedure.
20071005
Cell templates now adhere to the "NetworkReadyCell" syntax - fixed some minor bugs. Schmidt-Hieber.
20170328
* Added a new cell (cell_9) and an artificial cell (cell_10) that were not
part of the original 2007 publication.
* Added CA1 PC model (cell_11) for consistency. See
Golding NL, Mickus TJ, Katz Y, Kath WL, Spruston N (2005)
Factors mediating powerful voltage attenuation along CA1 pyramidal neuron
dendrites. J Physiol 568:69-82.
To run the original simulations as shown in the paper, use the original model from
https://www.janelia.org/sites/default/files/Attenuation.zip
Send bug reports and comments to
christoph.schmidt-hieber at uni-freiburg.de
Description of files:
./gc.hoc Starts the simulations. Requires mod files to be compiled.
./asin.mod arcsine function, forwarded to hoc from math.h
./spines.mod Introduces spine area scale factor as a range variable
(scale_spines).
Optionally, spine counts can be used as a range variable
as well (count_spines).
./synampa.mod Synaptic conductance change with a biexponential time course.
./share/calcSpines.hoc Calculate spine area scale factor from spine counts. Corrects
for spines hidden behind the shaft in 2D projections.
./share/config.hoc Simulation configuration settings.
./share/controlpanel.hoc Main control panel allowing cell and figure selection.
./share/figures.hoc Procedures reproducing figures.
./share/fixnseg.hoc d_lambda rule implementation.
./share/genutils.hoc Some general-purpose routines.
./share/spine.hoc Creates a spine, consisting of a neck (spine[0]) and a
head (spine[1])
./share/template.plt Template script file for creating a gnuplot script that
will show a 3D plot as in figure 6D.
./cell_x/membrane.hoc Loads cell morphology and connections, initializes spines,
inserts passive membrane properties. Uses a class "cell_x"
so that you can easily toggle between cells or use multiple
objects for network simulations.
./cell_x/morpho.hoc Loads morphology and spines without making use of a class.
Intended for standalone use of cells.
./cell_x/morpho.txt Coordinates and diameters of 3D points as plain text. Needed
by membrane.hoc to load 3D morphology from a procedure.
20071005
Cell templates now adhere to the "NetworkReadyCell" syntax - fixed some minor bugs. Schmidt-Hieber.
20170328
* Added a new cell (cell_9) and an artificial cell (cell_10) that were not
part of the original 2007 publication.
* Added CA1 PC model (cell_11) for consistency. See
Golding NL, Mickus TJ, Katz Y, Kath WL, Spruston N (2005)
Factors mediating powerful voltage attenuation along CA1 pyramidal neuron
dendrites. J Physiol 568:69-82.
To run the original simulations as shown in the paper, use the original model from
https://www.janelia.org/sites/default/files/Attenuation.zip