This is the ModelDB version of the readme for the model associated
with the paper:
Gurkiewicz M, Korngreen A (2007) A Numerical Approach to Ion Channel
Modelling Using Whole-Cell Voltage-Clamp Recordings and a Genetic
Algorithm PLoS Computational Biology 3(8):e169
Both the model (569 KB ZIP)
https://doi.org/10.1371/journal.pcbi.0030169.sd001
and the paper
http://dx.doi.org/10.1371/journal.pcbi.0030169
are available at the PLoS web site, and there is a copy of the
model also at ModelDB accession number 97756
In order to run the GA you have to install a recent version of NEURON
available from http://www.neuron.yale.edu
Once installed you can auto-launch from ModelDB and start the training algorithm (button press) or
Under linux:
------------
compile the KChannel.mod file using "nrnivmodl" then type
nrngui mosinit.hoc
Under Windows:
--------------
Run "mknrndll" then double click on the mosinit.hoc file icon.
Under MAC OS X:
---------------
On the MAC drag and drop the Ga_demo folder onto the mknrndll icon.
Then drag and drop the mosinit.hoc file onto the nrngui icon.
--------------
When the simulation starts a window with button choices will appear.
You can graph the performance of the best individual in the initial
starting population by clicking on the "Show best individual from
initial population" button. Click on the button labeled "Show result
of GA" to see a result of evolution of the model through 600
populations in the GA. Clicking on "Show final result of GA+praxis"
shows how the praxis method built into NEURON refined the GA result.
To run the entire demonstration training session click the "Start
training" button. When complete (after about 2 hours on a PC) you
will see matches for activation (the blue target curves are on top of
the red search results):
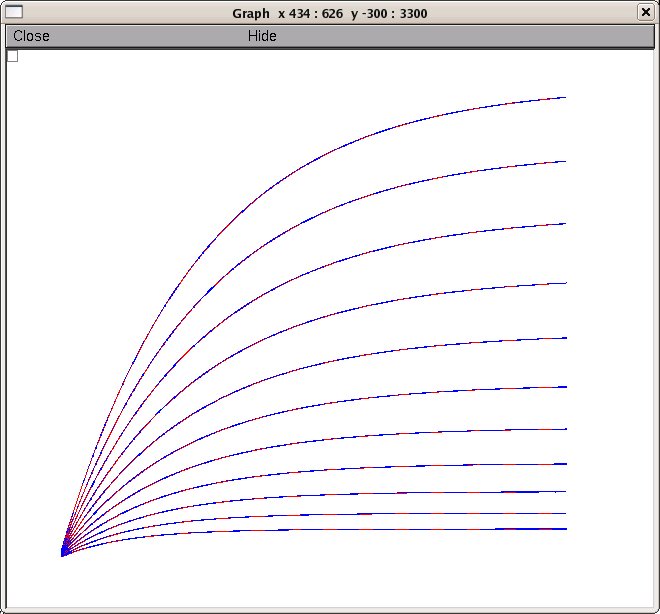 and deactivation
and deactivation
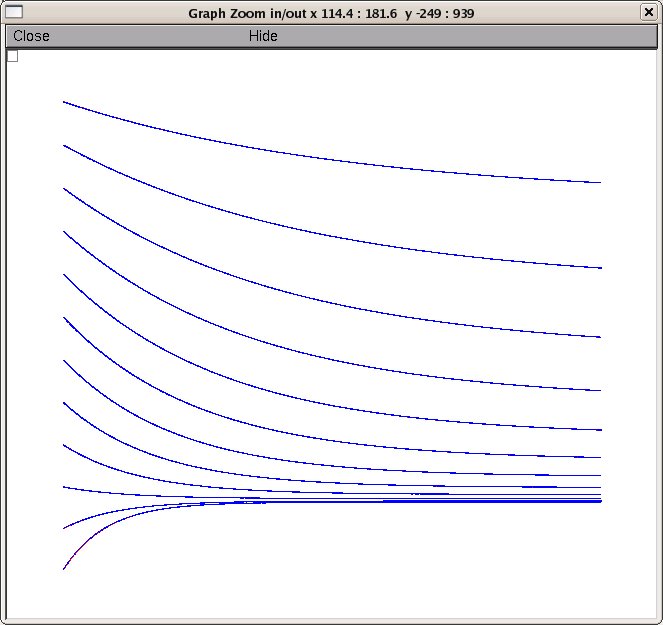
 and deactivation
and deactivation

and deactivation
