Frequently asked questions
Citations
How do I cite ModelDB?To cite ModelDB, we suggest the following recent paper:
McDougal RA, Morse TM, Carnevale T, Marenco L, Wang R, Migliore M, Miller PL, Shepherd GM, Hines ML. Twenty years of ModelDB and beyond: building essential modeling tools for the future of neuroscience. J Comput Neurosci. 2017; 42(1):1-10.
For examples when using code from ModelDB or sharing code on ModelDB, see our terms of use page, linked from the bottom of every ModelDB page.
If you use models from ModelDB, or information derived from those models, you should cite both ModelDB and the paper(s) that described the models. These papers appear under "reference(s):" in the model description page. Here is an example:
(after citing the papers that the models came from) ... "These network models used in the present study were obtained from ModelDB (McDougal et al, 2017; accession numbers 76883, 83514, and 83562)."
Sharing models
Can I share the source code for my submitted model publication with the reviewers? Can I make an unpublished or under review model publicly available?No. You can however use a read-only password (access code) to allow reviewers or collaborators see your model before it is made public. There is also a write access password that you can assign to share with collaborators.
Note: For extra help specific for NEURON models, click here.
- Create a directory that contains all the necessary model files and subdirectories.
- In the same directory, put a file (called readme.html, index.html, readme.txt, or README) that contains specific information and instructions on how to use the model. HTML is best because it allows formatting, images, links, and word-wrap. ModelDB will display this file by default in the Model Files window of the model description page.
-
Zip up the directory. On macOS (do not use the method of right-clicking to select Compress "Folder" because
it makes an archive which contains extra folders __MACOSX and files .DS_Store only used by the mac) and
unix/linux systems an appropriate terminal window command
for creating a zip file called ca1neuron.zip from a directory ca1nrn would be
zip -r ca1neuron.zip ca1nrn
On MSWin systems create your zip file by right clicking in the Windows Explorer dialog box on the background of the right part of the screen where your model files top level folder is displayed. Select "Send to" and then mouse off to the side to select "compressed (zipped) folder" from the pop-up menu. If desired, change the name of the newly created zip file making sure you retain the ".zip" file extension. - The zip file is now ready to be uploaded to ModelDB.
Step 0 (optional): Log in to ModelDB. This is not required, however it will make it easier for you to keep track of your models later.
Step 1: On the ModelDB homepage, click "Submit Model" to begin model submission.

Step 2: Enter your name and email address (so we can reach you), upload a zip file containing your model code, and (if not logged in) enter a password. Enter the citation information for your model in any format, if known. Note: only models with a valid citation can be made publicly available, however unpublished model codes may be shared with reviewers, collaborators, etc by entering a read access code.
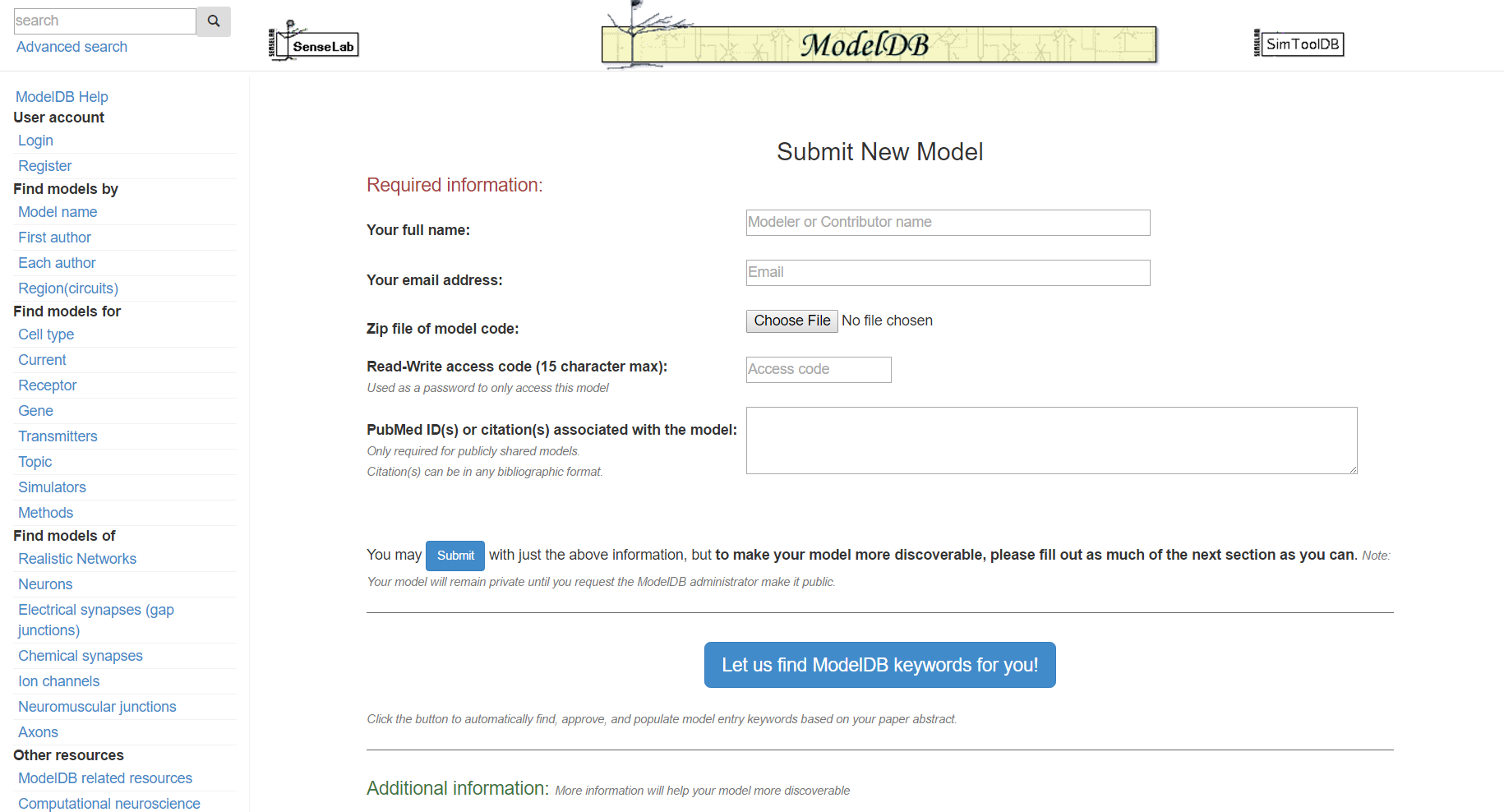
Step 3: Click "Let us find ModelDB keywords for you!" and paste in the abstract for the paper that goes with your model and/or enter additional keywords describing your model.
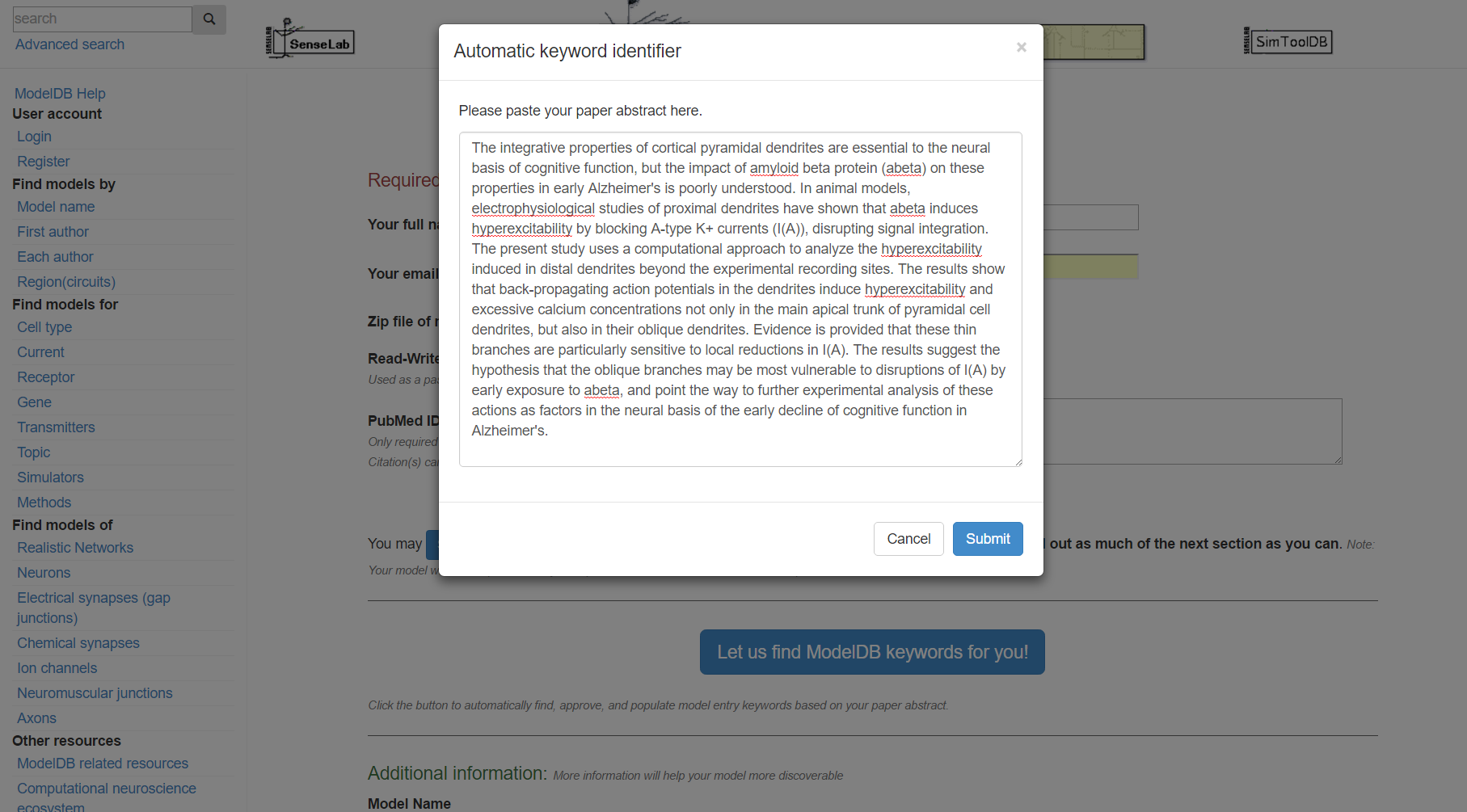
Step 4: Click "Submit", review the suggested metadata tags, deselect any that do not describe your model, and click "accept selected keywords".
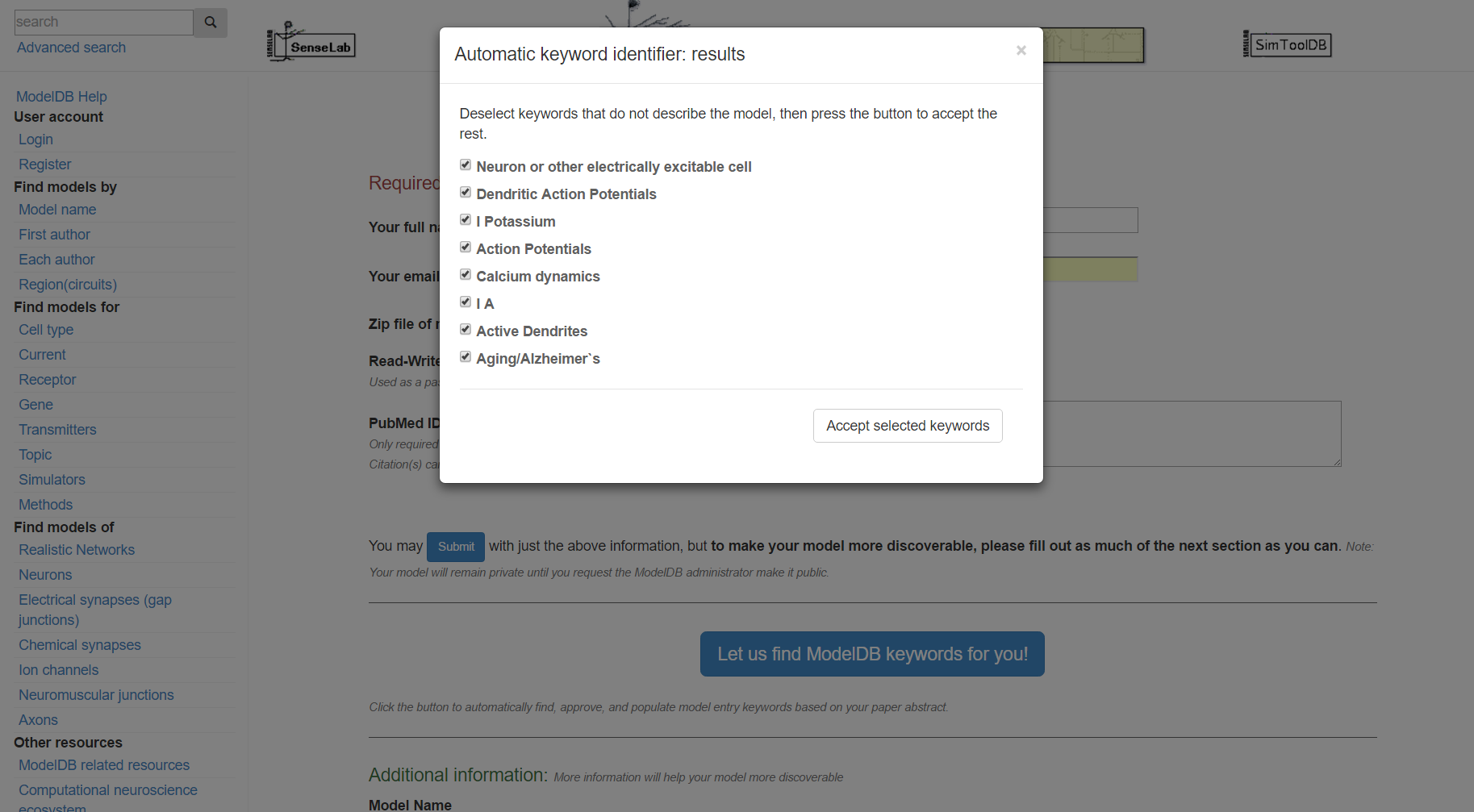
Step 5: Review the categorized metadata, add additional metadata (e.g. if your model has sodium channels but those were not previously suggested, annotate this at this time). This is not strictly required, however the more relevant metadata you include, the easier it will be for others to find your model. Click "submit" when finished. You will receive an email with links for accessing your model.
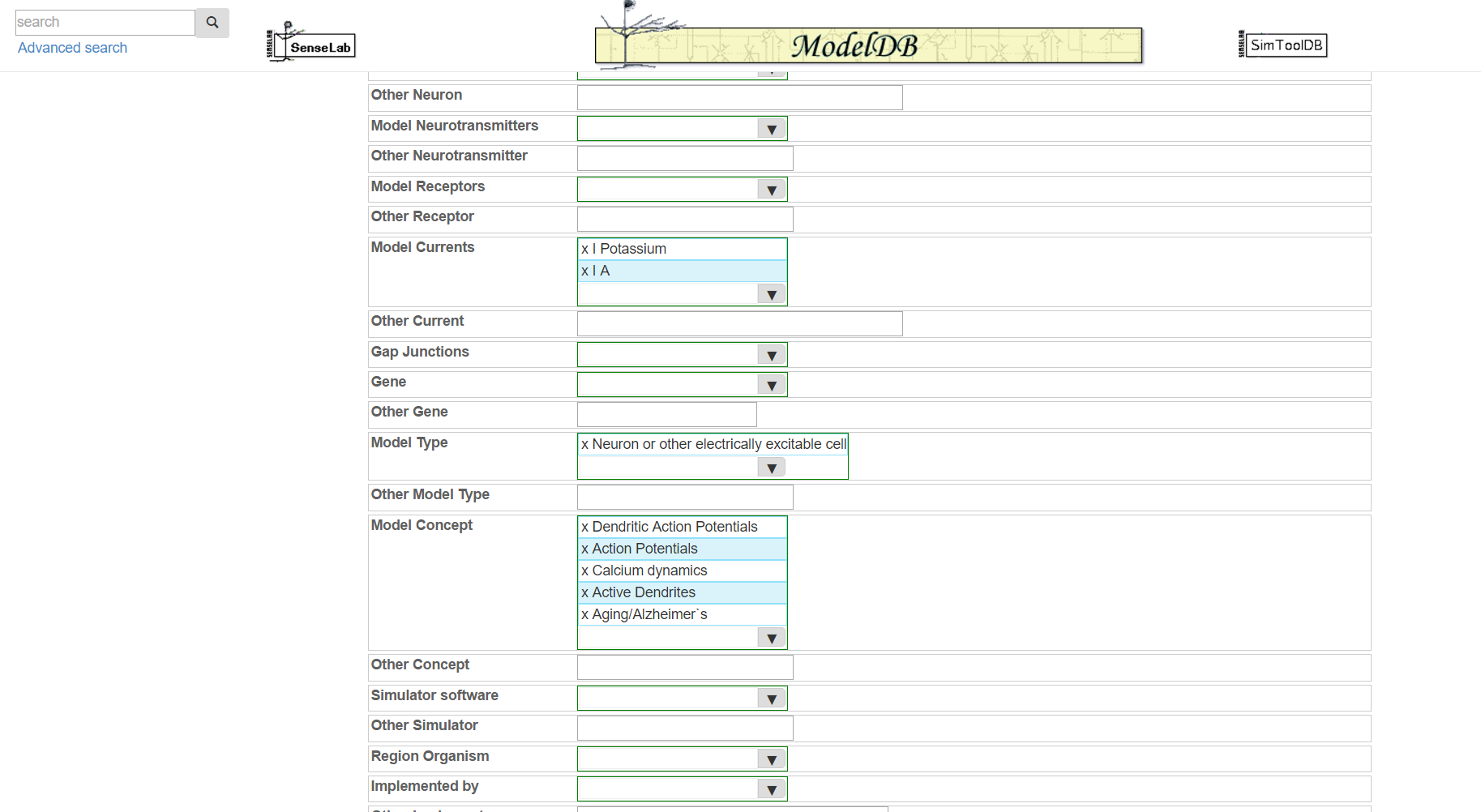
Step 6 (optional): Follow the prompts to provide additional information about your ion channel models for Ion Channel Genealogy.
Note: At this point, your model is not yet publicly accessible. If the accompanying paper is published, click "request to make public" on the model page to request review by a curator in preparation to make the model public. We may contact you if we have questions.
All models are created in private mode, and they remain that way until you request that they be made public. Do this by selecting "request to make public" on the model description page. A curator will then review your model and may contact you with questions before making it public.
All models are created in private mode, and they remain that way until you request that they be made public. Do this by selecting "request to make public" on the model description page. A curator will then review your model and may contact you with questions before making it public.
If the models were created while you were logged in, log in then click Show my private models on the navigation pane (on wide windows, this is displayed in the left column; otherwise navigation is available under the site menu).
The forgot submitted models page allows you to receive an email listing all private models associated with your email address. If you have forgotten the access code for a given model, you can click on the model's link in the email and then select "forgot access code". The next page will prompt you to reenter your email address; doing so and clicking submit will send you an email with a password reset link.
It is not possible to directly change a model after it is made public, due to the need to keep the public models curated. Instead, email the ModelDB Administrator (link in the footer) with the requested changes. A curator may contact you for additional information. Once accepted, the changes become the default version and all previous versions remain accessible via git.
Using ModelDB
What can I do with ModelDB?- Find a model described in a paper, download it, and experiment to understand the model's predictions.
- Find a model described in a paper. Use ModelView to understand the model's structure.
- Locate models and modeling papers on a given topic (e.g. Alzheimer's).
- Locate mod components (e.g. L-type calcium channels) for potential reuse.
- Search for simulator keywords (e.g. FInitializeHander) to find examples of how to use them.
- by accession number
- by a particular author
- by keyword (cell type, region, receptor, gene, transmitter, topic, simulator)
- use advanced search for ion currents: because these are short they are problematic to search with free text
- use advanced search for a combined keyword and full text search
- prefix case sensitive words with ^
- use * for completions
- use year: to search for models from given year; e.g. year:2015
- use file: to restrict results to a given filename; e.g. calcium file:*.mod
The advanced search link under the search box allows searching via both full text and attributes simultaneously.
Searching for papers by author instead of models is also supported; to do so, go here.
All locally stored models can be downloaded as a single file by pressing the "Download zip file" button. Some models can be run directly from the web page. For these models there will be a button labeled "Auto-launch".
ModelDB models are associated with many simulators and simulation environments. For some models the code is supplied externally and in these cases you will need to follow the links to the web sites where the code is made available. If you have any trouble finding the code at these external sites please email the ModelDB administrator which model (accession number) you were unable to find. For models supplied internally in ModelDB, when you press the Download button, depending on your browser's settings, the file will be downloaded to a previously choosen location (typically "Downloads" folder) or you will be asked to save the file in a directory and, if you want, you can change the name of the file. Once the file is downloaded you can expand the zip file with the unzip program that comes with your operating system (double clicking the zip file in windows explorer (mswin) or a finder window (mac) will usually automatically invoke an unzip program). In general you will need to install a simulator or simulation environment. Links below provide additional instructions for some simulators used by ModelDB models:
WARNING:
Running models on your machine is inherently insecure since models are able to execute any program or script consistent with the permissions of the user. We make a good faith effort at ModelDB to publicly provide models that are associated with published papers and do not knowingly provide models containing malicious or destructive code, viruses, worms, etc. We take several steps to ensure the safety of running our publicly available models. These include making a cursory examination of each models code, test-running each model, and not accepting anonymous models. However if we were "hacked" our verified files might be replaced with malicious ones. Therefore please inform us if running a model causes any kind of problem on your machine so that we may remove the model as soon as possible from the database.
If you think you've found an error in a model available through ModelDB, please contact the ModelDB administrator (link in footer) with a description of the issue. Possible errors include but are not limited to: logic errors, flipped signs, missing parentheses, etc. Note that every model is by definition an approximation, so missing ion channels, channels with the wrong kinetics, non-physiological parameter values are generally not considered modeling errors.
Many models have a "Model Views" tab at the top of their model description page. Click that tab, then select a figure under "Load Model Views". Two or more floating containers will appear, one with an expandable tree, the other(s) with graphical representations of the neuron(s). The tree may be expanded to see more information; e.g. on the view for "Figure 3" of model 87284 (Morse et al., 2010), clicking "1 cell with morphology", "root soma", "18 inserted mechanisms", "kad (kadist.mod)", "gkabar" will show the distribution of A type potassium currents governed by the kinetics in kadist.mod.
At the moment, ModelView only supports some NEURON and NeuroML models with a small number of cells. If the model you're interested in meets these criteria but still does not have a ModelView available, we may simply not have enabled it yet. Email the ModelDB administrator (link in footer) to request that we enable ModelView on a specific model.
- McDougal RA, Morse TM, Carnevale T, Marenco L, Wang R, Migliore M, Miller PL, Shepherd, GM, Hines ML (2017). Twenty years of ModelDB and beyond: building essential modeling tools for the future of neuroscience. Journal of Computational Neuroscience, 42(1), 1-10.
- Migliore M, Carnevale NT, Shepherd GM (2004) ModelDB: A Database to Support Computational Neuroscience. J Comput Neurosci 17:7-11
- Migliore M, Morse TM, Davison, AP, Marenco L, Shepherd GM, Hines ML. (2003) ModelDB: Making models publicly accessible to support computational neuroscience. Neuroinformatics 1:135-139.
- McDougal RA, Morse TM, Hines ML, Shepherd GM (2015). ModelView for ModelDB: online presentation of model structure. Neuroinformatics, 13(4), 459-470.
- McDougal RA, Dalal I, Morse TM, Shepherd GM (2019). Automated Metadata Suggestion During Repository Submission. Neuroinformatics.